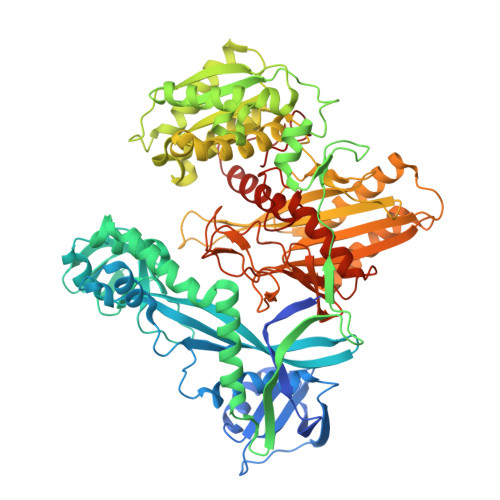
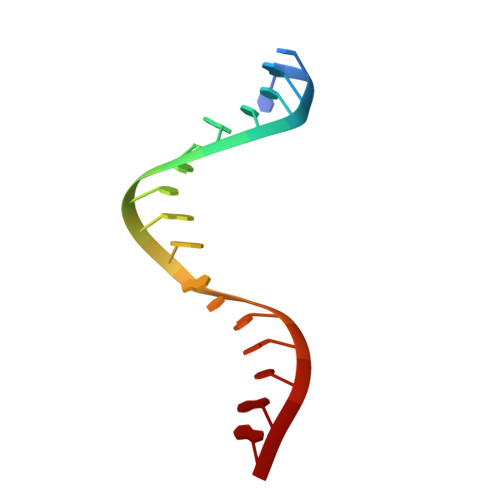
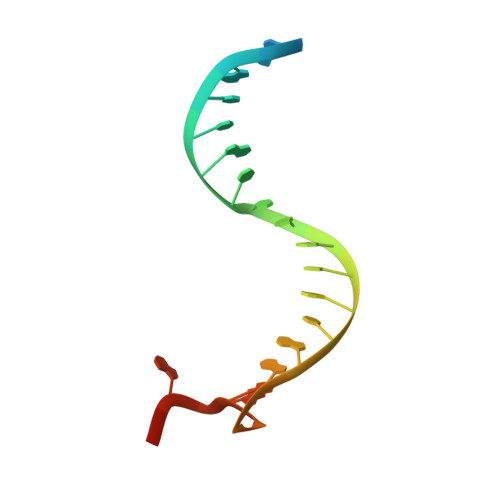
Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Liu, Y., Esyunina, D., Olovnikov, I., Teplova, M., Kulbachinskiy, A., Aravin, A.A., Patel, D.J.(2018) Cell Rep 24: 453-462
- PubMed: 29996105
- DOI: https://doi.org/10.1016/j.celrep.2018.06.021
- Primary Citation of Related Structures:
6D8A, 6D8F, 6D8P, 6D92, 6D95, 6D9K, 6D9L - PubMed Abstract:
Prokaryotic Argonaute (Ago) proteins were recently shown to target foreign genetic elements, thus making them a perfect model for studies of interference mechanisms. Here, we study interactions of Rhodobacter sphaeroides Ago (RsAgo) with guide RNA (gRNA) and fully complementary or imperfect target DNA (tDNA) using biochemical and structural approaches. We show that RsAgo can specifically recognize both the first nucleotide in gRNA and complementary nucleotide in tDNA, and both interactions contribute to nucleic acid binding. Non-canonical pairs and bulges on the target strand can be accommodated by RsAgo with minimal perturbation of the duplex but significantly reduce RsAgo affinity to tDNA. Surprisingly, mismatches between gRNA and tDNA induce dissociation of the guide-target duplex from RsAgo. Our results reveal plasticity in the ability of Ago proteins to accommodate helical imperfections, show how this might affect the efficiency of RNA silencing, and suggest a potential mechanism for guide release and Ago recycling.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: