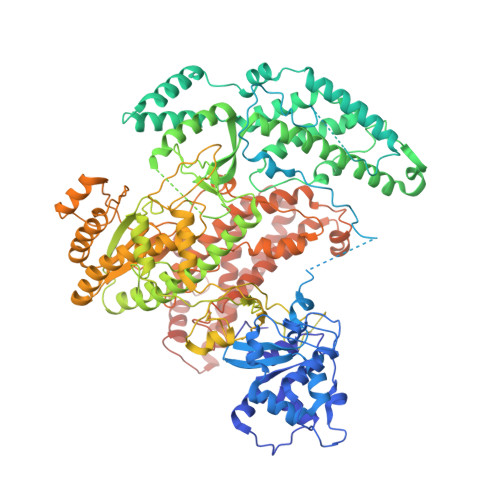
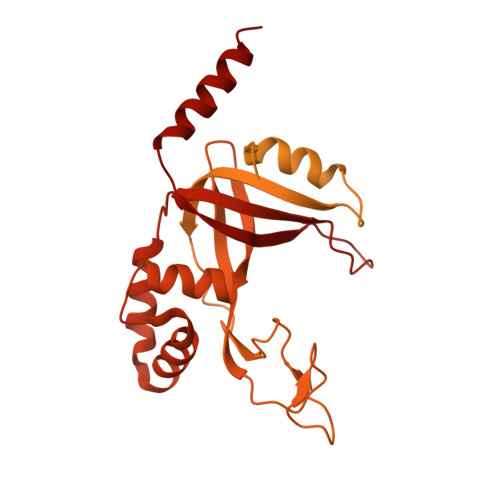
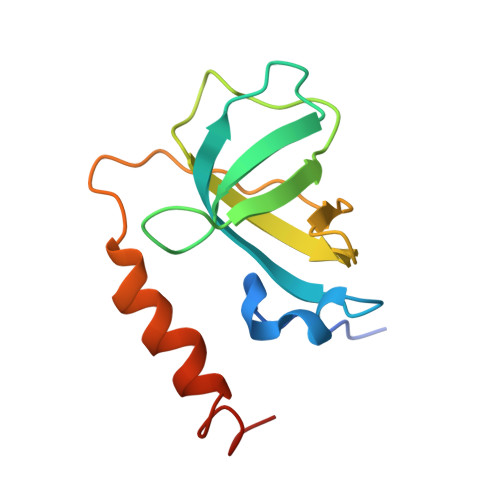
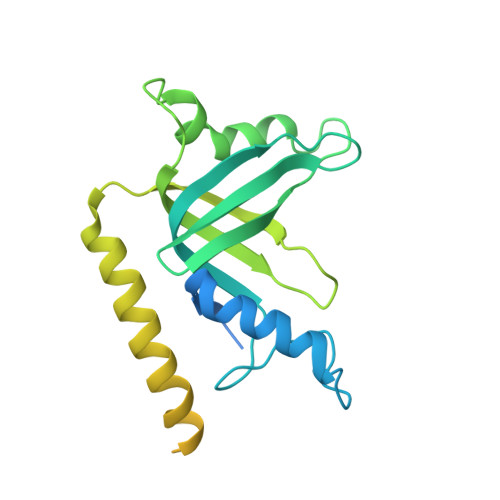
Structure of Telomerase with Telomeric DNA.
Jiang, J., Wang, Y., Susac, L., Chan, H., Basu, R., Zhou, Z.H., Feigon, J.(2018) Cell 173: 1179-1190.e13
- PubMed: 29775593
- DOI: https://doi.org/10.1016/j.cell.2018.04.038
- Primary Citation of Related Structures:
6D6V - PubMed Abstract:
Telomerase is an RNA-protein complex (RNP) that extends telomeric DNA at the 3' ends of chromosomes using its telomerase reverse transcriptase (TERT) and integral template-containing telomerase RNA (TER). Its activity is a critical determinant of human health, affecting aging, cancer, and stem cell renewal. Lack of atomic models of telomerase, particularly one with DNA bound, has limited our mechanistic understanding of telomeric DNA repeat synthesis. We report the 4.8 Å resolution cryoelectron microscopy structure of active Tetrahymena telomerase bound to telomeric DNA. The catalytic core is an intricately interlocked structure of TERT and TER, including a previously structurally uncharacterized TERT domain that interacts with the TEN domain to physically enclose TER and regulate activity. This complete structure of a telomerase catalytic core and its interactions with telomeric DNA from the template to telomere-interacting p50-TEB complex provides unanticipated insights into telomerase assembly and catalytic cycle and a new paradigm for a reverse transcriptase RNP.
- Department of Chemistry and Biochemistry, University of California, Los Angeles, Los Angeles, CA 90095, USA; Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, Los Angeles, CA 90095, USA; California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Organizational Affiliation: