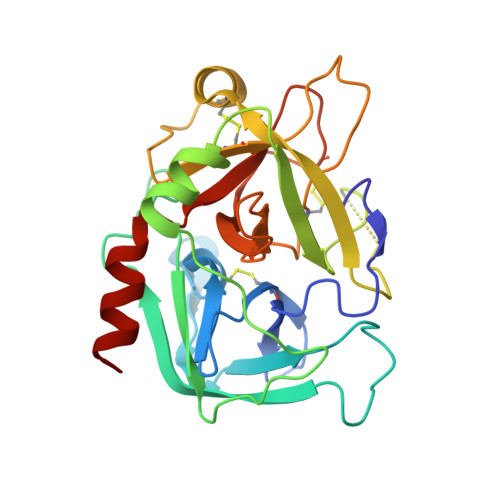
Reversible covalent direct thrombin inhibitors.
Sivaraja, M., Pozzi, N., Rienzo, M., Lin, K., Shiau, T.P., Clemens, D.M., Igoudin, L., Zalicki, P., Chang, S.S., Estiarte, M.A., Short, K.M., Williams, D.C., Datta, A., Di Cera, E., Kita, D.B.(2018) PLoS One 13: e0201377-e0201377
- PubMed: 30071045
- DOI: https://doi.org/10.1371/journal.pone.0201377
- Primary Citation of Related Structures:
6CYM - PubMed Abstract:
In recent years, the traditional treatments for thrombotic diseases, heparin and warfarin, are increasingly being replaced by novel oral anticoagulants offering convenient dosing regimens, more predictable anticoagulant responses, and less frequent monitoring. However, these drugs can be contraindicated for some patients and, in particular, their bleeding liability remains high. We have developed a new class of direct thrombin inhibitors (VE-DTIs) and have utilized kinetics, biochemical, and X-ray structural studies to characterize the mechanism of action and in vitro pharmacology of an exemplary compound from this class, Compound 1. We demonstrate that Compound 1, an exemplary VE-DTI, acts through reversible covalent inhibition. Compound 1 inhibits thrombin by transiently acylating the active site S195 with high potency and significant selectivity over other trypsin-like serine proteases. The compound inhibits the binding of a peptide substrate with both clot-bound and free thrombin with nanomolar potency. Compound 1 is a low micromolar inhibitor of thrombin activity against endogenous substrates such as fibrinogen and a nanomolar inhibitor of the activation of protein C and thrombin-activatable fibrinolysis inhibitor. In the thrombin generation assay, Compound 1 inhibits thrombin generation with low micromolar potency but does not increase the lag time for thrombin formation. In addition, Compound 1 showed weak inhibition of clotting in PT and aPTT assays consistent with its distinctive profile in the thrombin generation assay. Compound 1, while maintaining strong potency comparable to the current DTIs, has a distinct mechanism of action which produces a differentiating pharmacological profile. Acting through reversible covalent inhibition, these direct thrombin inhibitors could lead to new anticoagulants with better combined efficacy and bleeding profiles.
- Verseon Corporation, Fremont, California, United States of America.
Organizational Affiliation: