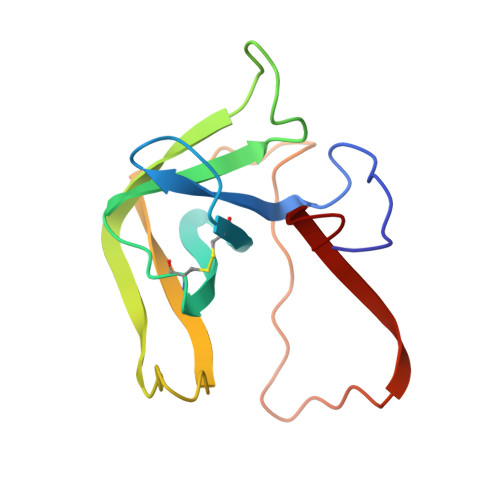
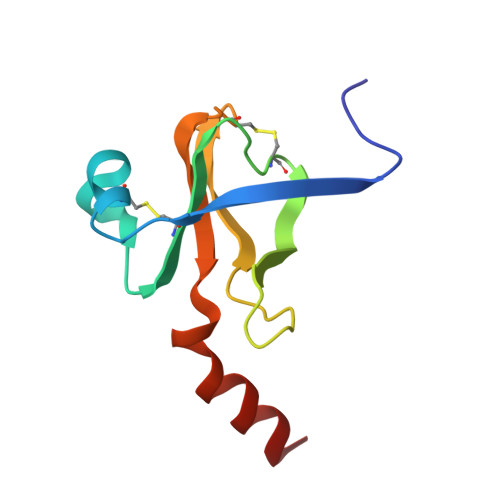
Structure of a tetrahedral transition state complex of alpha-chymotrypsin dimer at 1.8-A resolution.
Tulinsky, A., Blevins, R.A.(1987) J Biological Chem 262: 7737-7743
- PubMed: 3584139
- DOI: https://doi.org/10.2210/pdb6cha/pdb
- Primary Citation of Related Structures:
6CHA - PubMed Abstract:
A 1.8-A resolution x-ray crystallographic restrained least squares refinement has been carried out on the phenylethane boronic acid (PEBA) complex of alpha-chymotrypsin dimer (alpha-CHT), and it has been compared to the 1.67-A resolution structure of the native enzyme. PEBA has a high binding affinity for alpha-CHT, and the boronate forms a tetrahedral complex with Ser-195 OG of one molecule of the dimer; the boronate in the other molecule is severely disordered and does not form a tetrahedral complex. The former could be a model of the transition state of catalysis. The complex of PEBA X alpha-CHT displays significant nonequivalence in conformation of side chains between the independent molecules comparable to the native enzyme, but, like the latter, shows a high degree of fidelity in the folding of the main chain. The orientation of the phenyl ring, CA and CB of PEBA, in the specificity sites of the two molecules is similar, suggesting that recognition is fairly insensitive to small departures from local symmetry; the same does not apply to the boronate functionalities suggesting that greater precision is required for catalysis. The folding of the molecule remains the same upon PEBA binding, but some of the side chains respond nonequivalently. The latter is a consequence of the inherent nonequivalence of the native dimer and the asymmetrical nature of the PEBA binding.