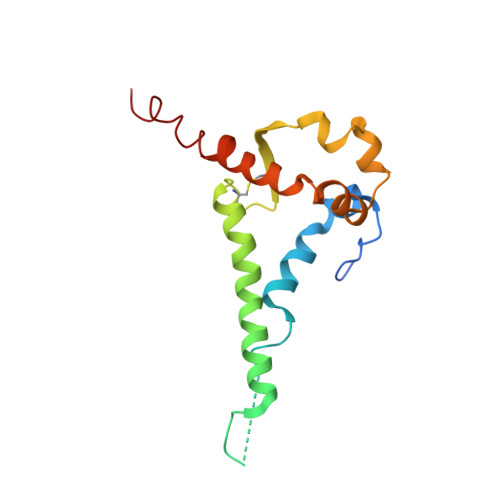
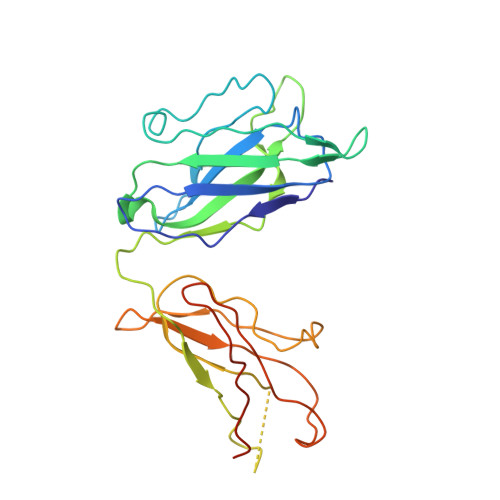
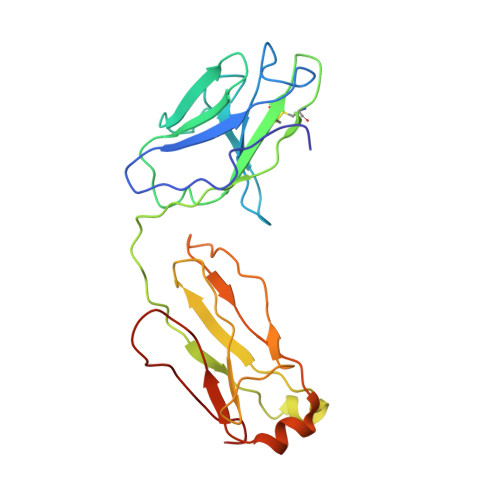
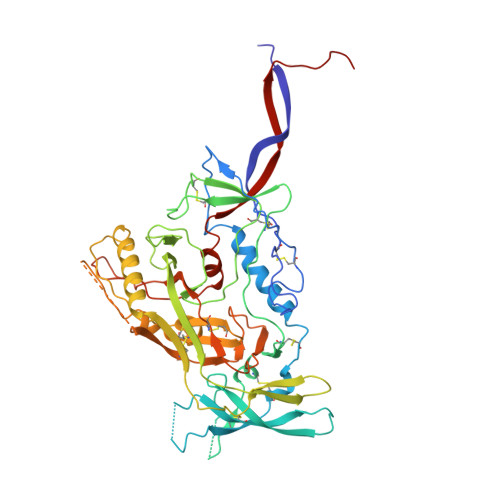
Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope.
Barnes, C.O., Gristick, H.B., Freund, N.T., Escolano, A., Lyubimov, A.Y., Hartweger, H., West, A.P., Cohen, A.E., Nussenzweig, M.C., Bjorkman, P.J.(2018) Nat Commun 9: 1251-1251
- PubMed: 29593217
- DOI: https://doi.org/10.1038/s41467-018-03632-y
- Primary Citation of Related Structures:
6CH7, 6CH8, 6CH9, 6CHB - PubMed Abstract:
Broadly neutralizing antibodies (bNAbs) isolated from HIV-1-infected individuals inform HIV-1 vaccine design efforts. Developing bNAbs with increased efficacy requires understanding how antibodies interact with the native oligomannose and complex-type N-glycan shield that hides most protein epitopes on HIV-1 envelope (Env). Here we present crystal structures, including a 3.8-Å X-ray free electron laser dataset, of natively glycosylated Env trimers complexed with BG18, the most potent V3/N332 gp120 glycan-targeting bNAb reported to date. Our structures show conserved contacts mediated by common D gene-encoded residues with the N332 gp120 glycan and the gp120 GDIR peptide motif, but a distinct Env-binding orientation relative to PGT121/10-1074 bNAbs. BG18's binding orientation provides additional contacts with N392 gp120 and N386 gp120 glycans near the V3-loop base and engages protein components of the V1-loop. The BG18-natively-glycosylated Env structures facilitate understanding of bNAb-glycan interactions critical for using V3/N332 gp120 bNAbs therapeutically and targeting their epitope for immunogen design.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA, 91125, USA.
Organizational Affiliation: