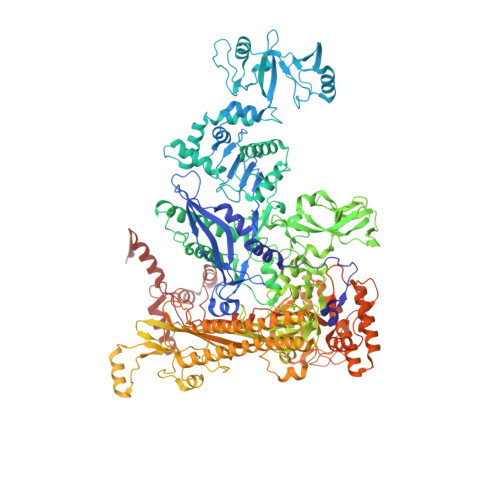
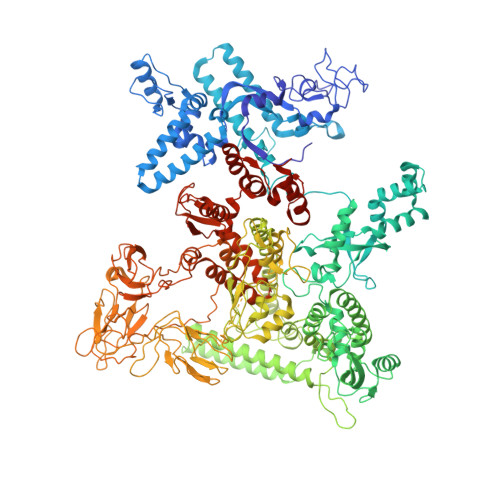
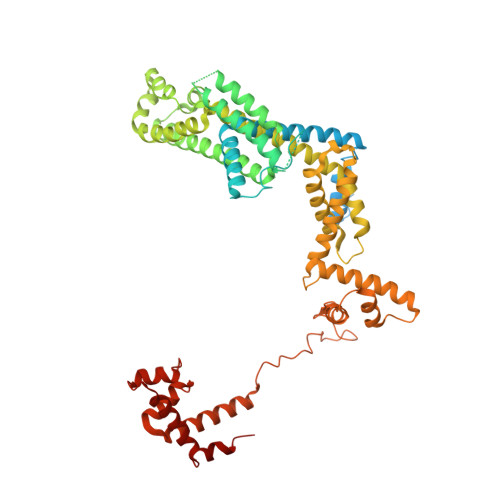
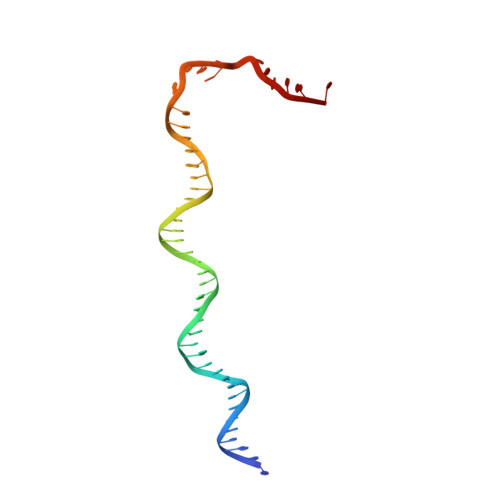
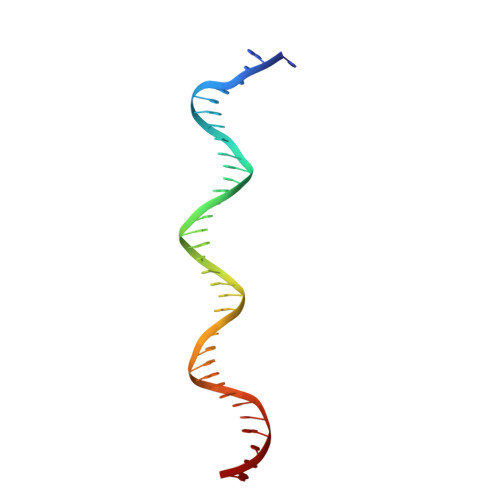
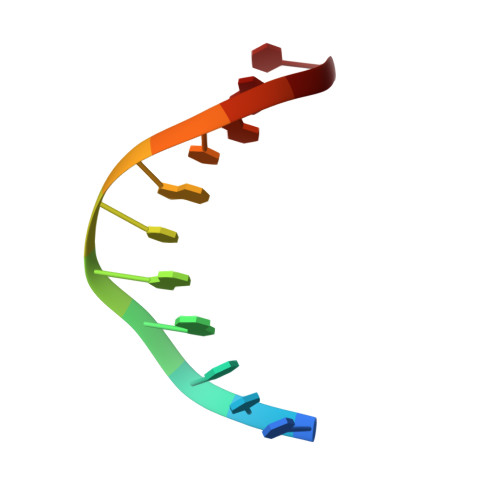
Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation.
Narayanan, A., Vago, F.S., Li, K., Qayyum, M.Z., Yernool, D., Jiang, W., Murakami, K.S.(2018) J Biological Chem 293: 7367-7375
- PubMed: 29581236
- DOI: https://doi.org/10.1074/jbc.RA118.002161
- Primary Citation of Related Structures:
6C9Y, 6CA0 - PubMed Abstract:
First step of gene expression is transcribing the genetic information stored in DNA to RNA by the transcription machinery including RNA polymerase (RNAP). In Escherichia coli , a primary σ 70 factor forms the RNAP holoenzyme to express housekeeping genes. The σ 70 contains a large insertion between the conserved regions 1.2 and 2.1, the σ non-conserved region (σ NCR ), but its function remains to be elucidated. In this study, we determined the cryo-EM structures of the E. coli RNAP σ 70 holoenzyme and its complex with promoter DNA (open complex, RPo) at 4.2 and 5.75 Å resolutions, respectively, to reveal native conformations of RNAP and DNA. The RPo structure presented here found an interaction between the σ NCR and promoter DNA just upstream of the -10 element, which was not observed in a previously determined E. coli RNAP transcription initiation complex (RPo plus short RNA) structure by X-ray crystallography because of restraint of crystal packing effects. Disruption of the σ NCR and DNA interaction by the amino acid substitutions (R157A/R157E) influences the DNA opening around the transcription start site and therefore decreases the transcription activity of RNAP. We propose that the σ NCR and DNA interaction is conserved in proteobacteria, and RNAP in other bacteria replaces its role with a transcription factor.
- Department of Biochemistry and Molecular Biology, The Center for RNA Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802; Department of Biological Sciences, Markey Center for Structural Biology, Purdue University, West Lafayette, Indiana 47906.
Organizational Affiliation: