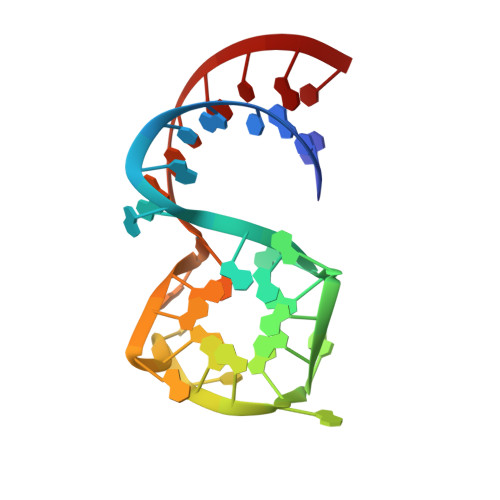
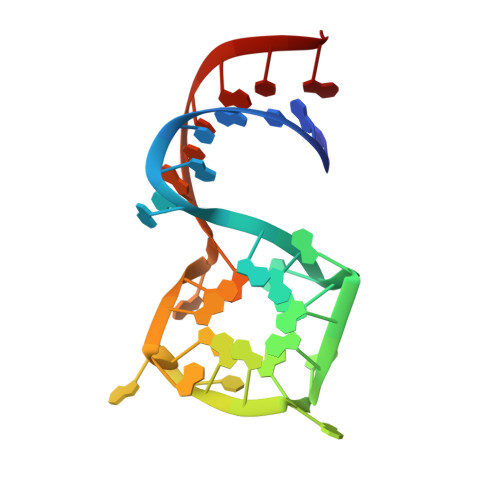
Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Trachman 3rd., R.J., Abdolahzadeh, A., Andreoni, A., Cojocaru, R., Knutson, J.R., Ryckelynck, M., Unrau, P.J., Ferre-D'Amare, A.R.(2018) Biochemistry 57: 3544-3548
- PubMed: 29768001
- DOI: https://doi.org/10.1021/acs.biochem.8b00399
- Primary Citation of Related Structures:
6C63, 6C64, 6C65 - PubMed Abstract:
Several RNA aptamers that bind small molecules and enhance their fluorescence have been successfully used to tag and track RNAs in vivo, but these genetically encodable tags have not yet achieved single-fluorophore resolution. Recently, Mango-II, an RNA that binds TO1-Biotin with ∼1 nM affinity and enhances its fluorescence by >1500-fold, was isolated by fluorescence selection from the pool that yielded the original RNA Mango. We determined the crystal structures of Mango-II in complex with two fluorophores, TO1-Biotin and TO3-Biotin, and found that despite their high affinity, the ligands adopt multiple distinct conformations, indicative of a binding pocket with modest stereoselectivity. Mutational analysis of the binding site led to Mango-II(A22U), which retains high affinity for TO1-Biotin but now discriminates >5-fold against TO3-biotin. Moreover, fluorescence enhancement of TO1-Biotin increases by 18%, while that of TO3-Biotin decreases by 25%. Crystallographic, spectroscopic, and analogue studies show that the A22U mutation improves conformational homogeneity and shape complementarity of the fluorophore-RNA interface. Our work demonstrates that even after extensive functional selection, aptamer RNAs can be further improved through structure-guided engineering.
- Biochemistry and Biophysics Center , National Heart, Lung and Blood Institute , 50 South Drive, MSC 8012 , Bethesda , Maryland 20892-8012 , United States.
Organizational Affiliation: