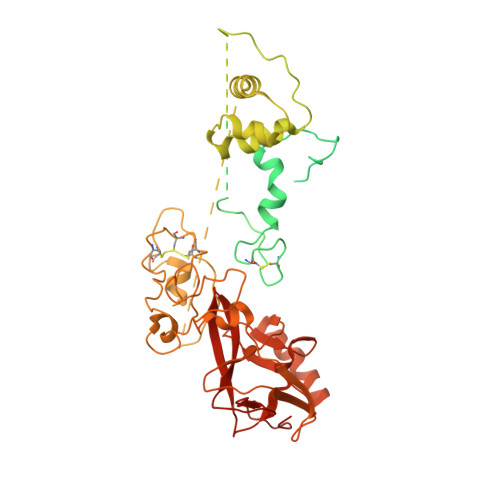
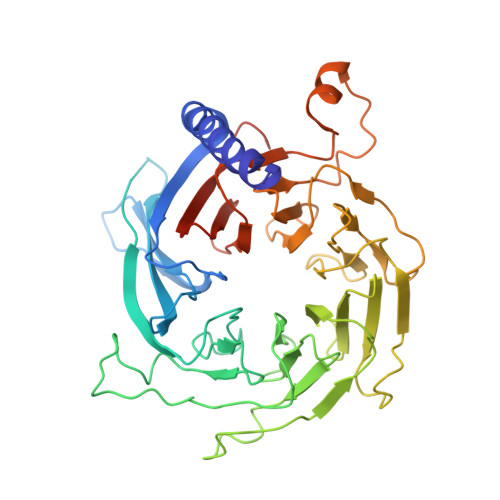
Structures of human PRC2 with its cofactors AEBP2 and JARID2.
Kasinath, V., Faini, M., Poepsel, S., Reif, D., Feng, X.A., Stjepanovic, G., Aebersold, R., Nogales, E.(2018) Science 359: 940-944
- PubMed: 29348366
- DOI: https://doi.org/10.1126/science.aar5700
- Primary Citation of Related Structures:
6C23, 6C24 - PubMed Abstract:
Transcriptionally repressive histone H3 lysine 27 methylation by Polycomb repressive complex 2 (PRC2) is essential for cellular differentiation and development. Here we report cryo-electron microscopy structures of human PRC2 in a basal state and two distinct active states while in complex with its cofactors JARID2 and AEBP2. Both cofactors mimic the binding of histone H3 tails. JARID2, methylated by PRC2, mimics a methylated H3 tail to stimulate PRC2 activity, whereas AEBP2 interacts with the RBAP48 subunit, mimicking an unmodified H3 tail. SUZ12 interacts with all other subunits within the assembly and thus contributes to the stability of the complex. Our analysis defines the complete architecture of a functionally relevant PRC2 and provides a structural framework to understand its regulation by cofactors, histone tails, and RNA.
- QB3 Institute, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: