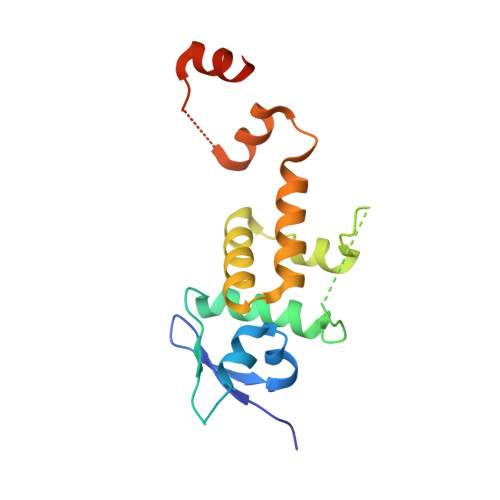
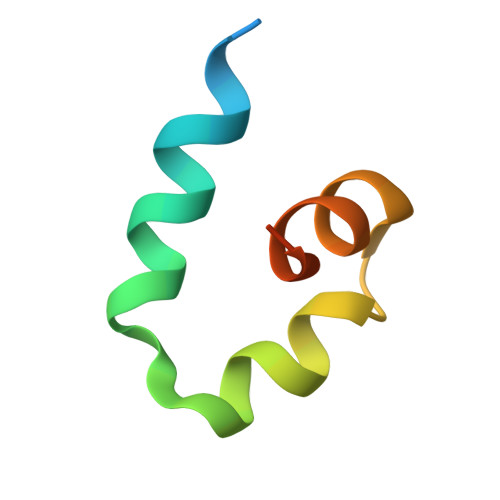
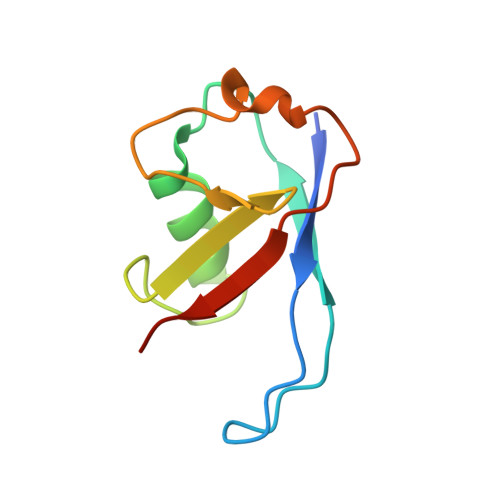
A Structure-Based Strategy for Engineering Selective Ubiquitin Variant Inhibitors of Skp1-Cul1-F-Box Ubiquitin Ligases.
Gorelik, M., Manczyk, N., Pavlenco, A., Kurinov, I., Sidhu, S.S., Sicheri, F.(2018) Structure 26: 1226
- PubMed: 30033217
- DOI: https://doi.org/10.1016/j.str.2018.06.004
- Primary Citation of Related Structures:
6BVA, 6BYH, 6C16 - PubMed Abstract:
Skp1-Cul1-F-box (SCF) E3 ligases constitute the largest and best-characterized family of the multisubunit E3 ligases with important cellular functions and numerous disease links. The specificity of an SCF E3 ligase is established by one of the 69 human F-box proteins that are recruited to Cul1 through the Skp1 adaptor. We previously reported generation of ubiquitin variants (UbVs) targeting Fbw7 and Fbw11, which inhibit ligase activity by binding at the F-box-Skp1 interface to competitively displace Cul1. In the present study, we employed an optimized engineering strategy to generate specific binding UbVs against 17 additional Skp1-F-box complexes. We validated our design strategy and uncovered the structural basis of binding specificity by crystallographic analyses of representative UbVs bound to Skp1-Fbl10 and Skp1-Fbl11. Our study highlights the power of combining phage display with structure-based design to develop UbVs targeting specific protein surfaces.
- Banting and Best Department of Medical Research, Terrence Donnelly Center for Cellular and Biomolecular Research, University of Toronto, Toronto, ON M5S 3E1, Canada; Department of Molecular Genetics, University of Toronto, Toronto, ON M5S 1A8, Canada.
Organizational Affiliation: