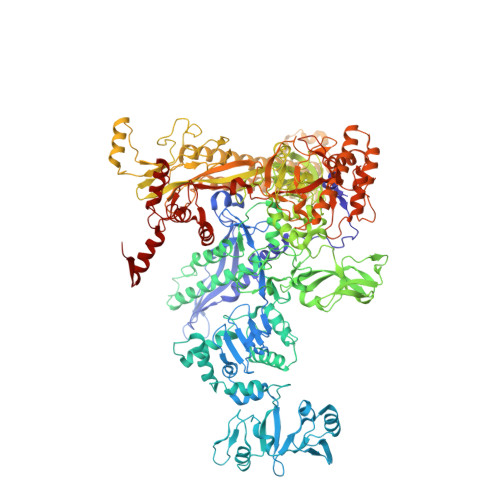
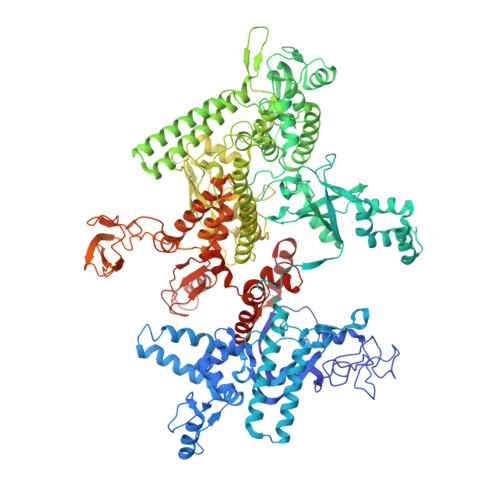
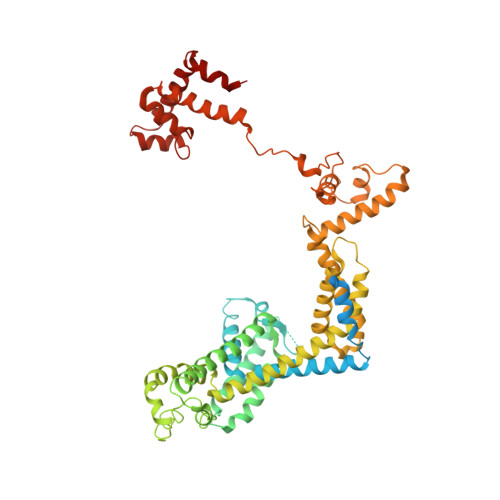
Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro.
Bruhn-Olszewska, B., Molodtsov, V., Sobala, M., Dylewski, M., Murakami, K.S., Cashel, M., Potrykus, K.(2018) Biochim Biophys Acta 1861: 731-742
- PubMed: 30012465
- DOI: https://doi.org/10.1016/j.bbagrm.2018.07.005
- Primary Citation of Related Structures:
6BYU - PubMed Abstract:
Precise regulation of gene expression is crucial for bacteria to respond to changing environmental conditions. In addition to protein factors affecting RNA polymerase (RNAP) activity, second messengers play an important role in transcription regulation, such as well-known effectors of the stringent response: guanosine 5'triphosphate-3'diphosphate and guanosine 3', 5'-bis(diphosphate) [(p)ppGpp]. Although much is known about importance of the 5' and 3' moieties of (p)ppGpp, the role of the guanine base remains somewhat cryptic. Here, we use (p)ppGpp's adenine analogs [(p)ppApp] to investigate how the nucleobase contributes to determine its binding site and transcriptional regulation. We determined X-ray crystal structure of Escherichia coli RNAP-(p)ppApp complex, which shows the analogs bind near the active site and switch regions of RNAP. We have also explored the regulatory effects of (p)ppApp on transcription initiating from the well-studied E. coli rrnB P1 promoter to assess and compare properties of (p)ppApp with (p)ppGpp. We demonstrate that contrary to (p)ppGpp, (p)ppApp activates transcription at this promoter and DksA hinders this effect. Moreover, pppApp exerts a stronger effect than ppApp. We also show that when ppGpp and pppApp are present together, the outcome depends on which one of them was pre-incubated with RNAP first. This behavior suggests a surprising Yin-Yang like reciprocal plasticity of RNAP responses at a single promoter, occasioned simply by pre-exposure to one or the other nucleotide. Our observations underscore the importance of the (p)ppNpp's purine nucleobase for interactions with RNAP, which may lead to a better fundamental understanding of (p)ppGpp regulation of RNAP activity.
- Department of Bacterial Molecular Genetics, Faculty of Biology, University of Gdansk, Wita Stwosza 59, 80-308 Gdansk, Poland. Electronic address: b.bruhn.olszewska@ug.edu.pl.
Organizational Affiliation: