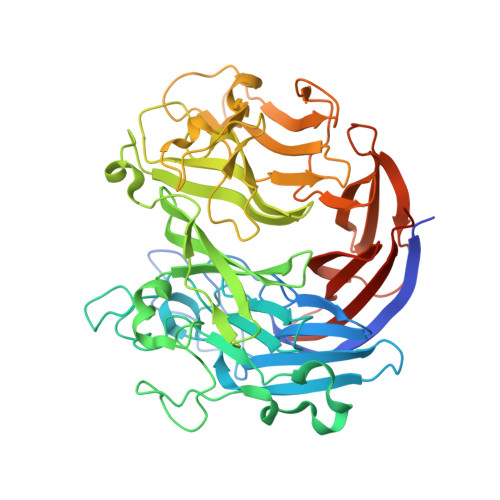
Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
Ulaganathan, T., Helbert, W., Kopel, M., Banin, E., Cygler, M.(2018) J Biological Chem 293: 4026-4036
- PubMed: 29382716
- DOI: https://doi.org/10.1074/jbc.RA117.001642
- Primary Citation of Related Structures:
6BYP, 6BYT, 6BYX - PubMed Abstract:
Ulvan is a major cell wall component of green algae of the genus Ulva , and some marine bacteria encode enzymes that can degrade this polysaccharide. The first ulvan-degrading lyases have been recently characterized, and several putative ulvan lyases have been recombinantly expressed, confirmed as ulvan lyases, and partially characterized. Two families of ulvan-degrading lyases, PL24 and PL25, have recently been established. The PL24 lyase LOR_107 from the bacterial Alteromonadales sp. strain LOR degrades ulvan endolytically, cleaving the bond at the C4 of a glucuronic acid. However, the mechanism and LOR_107 structural features involved are unknown. We present here the crystal structure of LOR_107, representing the first PL24 family structure. We found that LOR_107 adopts a seven-bladed β-propeller fold with a deep canyon on one side of the protein. Comparative sequence analysis revealed a cluster of conserved residues within this canyon, and site-directed mutagenesis disclosed several residues essential for catalysis. We also found that LOR_107 uses the His/Tyr catalytic mechanism, common to several PL families. We captured a tetrasaccharide substrate in the structures of two inactive mutants, which indicated a two-step binding event, with the first substrate interaction near the top of the canyon coordinated by Arg 320 , followed by sliding of the substrate into the canyon toward the active-site residues. Surprisingly, the LOR_107 structure was very similar to that of the PL25 family PLSV_3936, despite only ∼14% sequence identity between the two enzymes. On the basis of our structural and mutational analyses, we propose a catalytic mechanism for LOR_107 that differs from the typical His/Tyr mechanism.
- From the Department of Biochemistry, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada.
Organizational Affiliation: