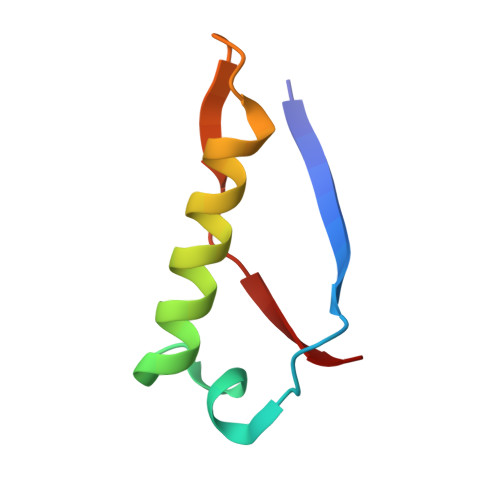
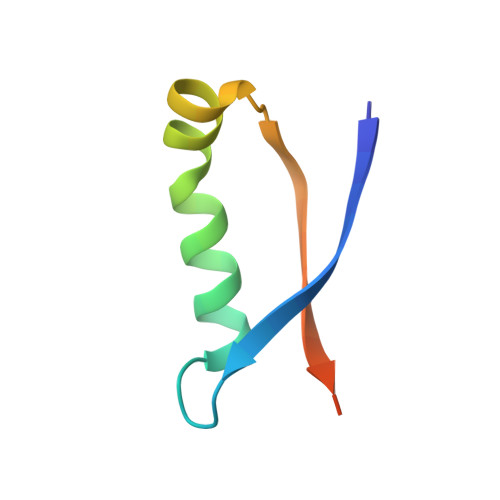
High-resolution structure of intramolecularly proteolyzed human mucin-1 SEA domain.
Noguera, M.E., Jakoncic, J., Ermacora, M.R.(2020) Biochim Biophys Acta Proteins Proteom 1868: 140361-140361
- PubMed: 31923589
- DOI: https://doi.org/10.1016/j.bbapap.2020.140361
- Primary Citation of Related Structures:
6BSB, 6BSC - PubMed Abstract:
SEA domains are ubiquitous in large proteins associated with highly glycosylated environments. Certain SEA domains undergo intramolecular proteolysis involving a nucleophilic attack of a serine hydroxyl group on the preceding glycine carbonyl. The mucin-1 (MUC1) SEA domain has been extensively investigated as a model of intramolecular proteolysis. Since neither a general base, a general acid, nor an oxyanion hole could be identified in MUC1 SEA, it has been suggested that proteolysis is accelerated by a non-planarity of the scissile peptide bond imposed by protein folding. A reactant distorted peptide bond has been also invoked to explain the autoproteolysis of several unrelated proteins. However, the only evidence of peptide distortion in MUC1 SEA stems from molecular dynamic simulations of the reactant modeled upon a single NMR structure of the cleaved product. We report the first high-resolution X-ray structure of cleaved MUC1 SEA. Structural comparison with uncleaved SEA domains suggests that the number of residues evolutionarily inserted in the cleaved loop of MUC1 SEA precludes the formation of a properly hydrogen-bonded beta turn. By sequence analysis, we show that this conformational frustration is shared by all known cleaved SEA domains. In addition, alternative conformations of the uncleaved precursor could be modeled in which the scissile peptide bond is planar. The implications of these structures for autoproteolysis are discussed in the light of the previous research on autoproteolysis.
- Departamento de Ciencia y Tecnología, Universidad Nacional de Quilmes, Argentina; Instituto de Química y Físico-Química Biológicas, Universidad de Buenos Aires, Buenos Aires, Argentina.
Organizational Affiliation: