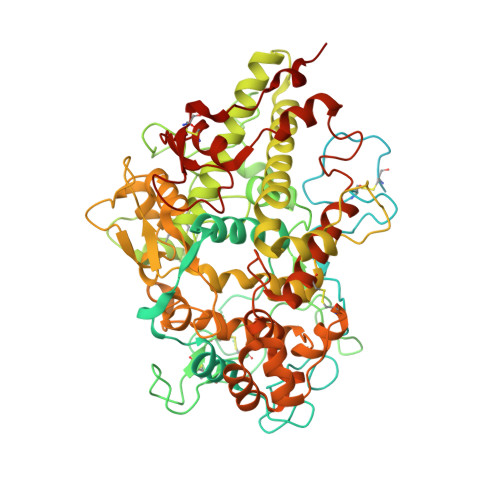
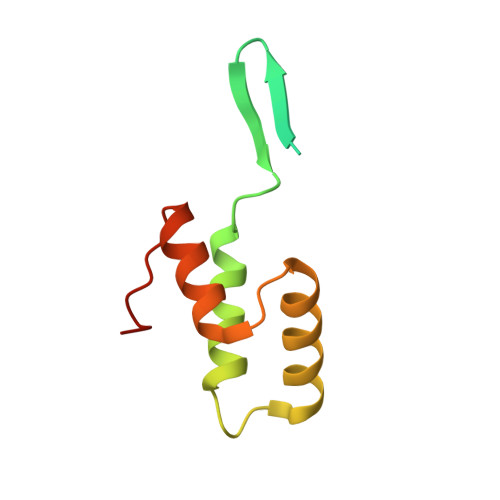
Identification and structural characterization of a novel myeloperoxidase inhibitor from Staphylococcus delphini.
Ploscariu, N.T., de Jong, N.W.M., van Kessel, K.P.M., van Strijp, J.A.G., Geisbrecht, B.V.(2018) Arch Biochem Biophys 645: 1-11
- PubMed: 29524428
- DOI: https://doi.org/10.1016/j.abb.2018.03.007
- Primary Citation of Related Structures:
6BMT - PubMed Abstract:
Staphylococcus aureus and related species are highly adapted to their hosts and have evolved numerous strategies to evade the immune system. S. aureus shows resistance to killing following uptake into the phagosome, which suggests that the bacterium evades intracellular killing mechanisms used by neutrophils. We recently discovered an S. aureus protein (SPIN for Staphylococcal Peroxidase INhibitor) that binds to and inhibits myeloperoxidase (MPO), a major player in the oxidative defense of neutrophils. To allow for comparative studies between multiple SPIN sequences, we identified a panel of homologs from species closely related to S. aureus. Characterization of these proteins revealed that SPIN molecules from S. agnetis, S. delphini, S. schleiferi, and S. intermedius all bind human MPO with nanomolar affinities, and that those from S. delphini, S. schleiferi, and S. intermedius inhibit human MPO in a dose-dependent manner. A 2.4 Å resolution co-crystal structure of SPIN-delphini bound to recombinant human MPO allowed us to identify conserved structural features of SPIN proteins, and to propose sequence-dependent physical explanations for why SPIN-aureus binds human MPO with higher affinity than SPIN-delphini. Together, these studies expand our understanding of MPO binding and inhibition by a recently identified component of the staphylococcal innate immune evasion arsenal.
- Department of Biochemistry & Molecular Biophysics, Kansas State University, 141 Chalmers Hall, 1711 Claflin Road, Manhattan, KS, 66506, USA.
Organizational Affiliation: