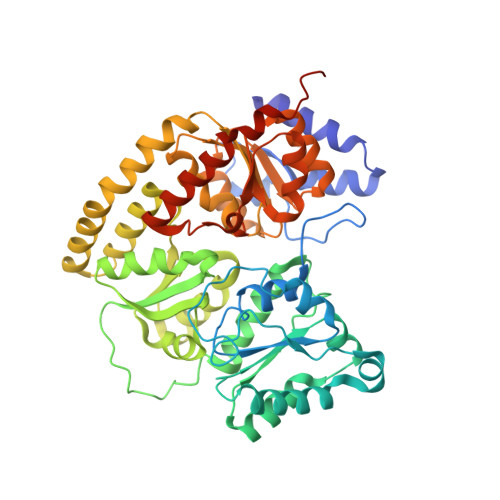
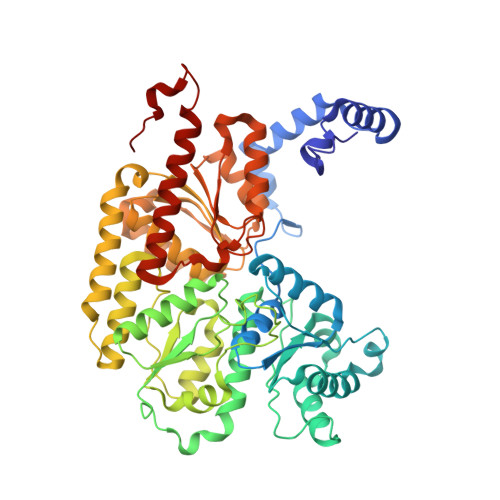
Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
Keable, S.M., Vertemara, J., Zadvornyy, O.A., Eilers, B.J., Danyal, K., Rasmussen, A.J., De Gioia, L., Zampella, G., Seefeldt, L.C., Peters, J.W.(2017) J Inorg Biochem 180: 129-134
- PubMed: 29275221
- DOI: https://doi.org/10.1016/j.jinorgbio.2017.12.008
- Primary Citation of Related Structures:
6BBL - PubMed Abstract:
The biological reduction of dinitrogen (N 2 ) to ammonia is catalyzed by the complex metalloenzyme nitrogenase. Structures of the nitrogenase component proteins, Iron (Fe) protein and Molybdenum‑iron (MoFe) protein, and the stabilized complexes these component proteins, have been determined, providing a foundation for a number of fundamental aspects of the complicated catalytic mechanism. The reduction of dinitrogen to ammonia is a complex process that involves the binding of N 2 followed by reduction with multiple electrons and protons. Electron transfer into nitrogenase is typically constrained to the unique electron donor, the Fe protein. These constraints have prevented structural characterization of the active site with bound substrate. Recently it has been realized that selected amino acid substitutions in the environment of the active site metal cluster (Iron‑molybdenum cofactor, FeMo-co) allow substrates to persist even in the resting state. Reported here is a 1.70Å crystal structure of a nitrogenase MoFe protein α-96 Arg➔Gln variant with the alternative substrate acetylene trapped in a channel in close proximity to FeMo-co. Complementary theoretical calculations support the validity of the acetylene interaction at this site and is also consistent with more favorable interactions in the variant MoFe protein compared to the native MoFe protein. This work represents the first structural evidence of a substrate trapped in the nitrogenase MoFe protein and is consistent with earlier assignments of proposed substrate pathways and substrate binding sites deduced from biochemical, spectroscopic, and theoretical studies.
- Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT 59717, United States.
Organizational Affiliation: