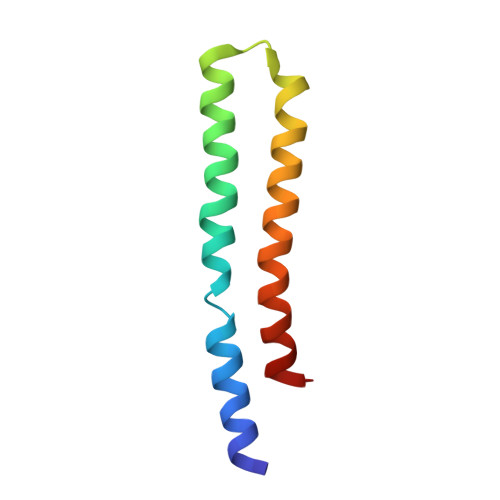
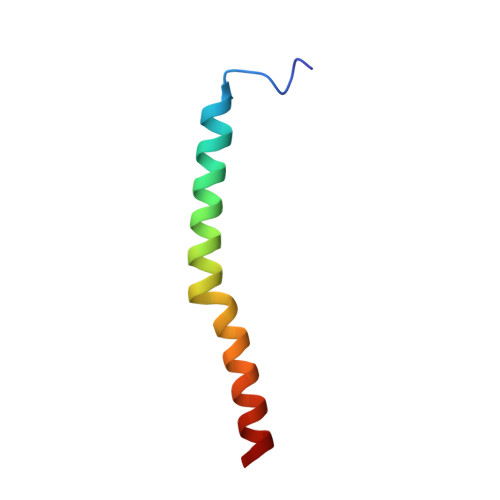
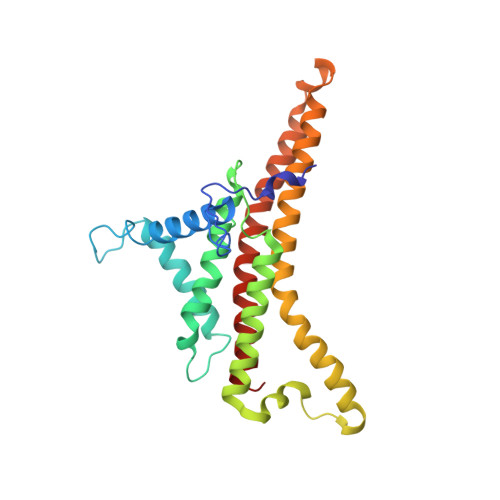
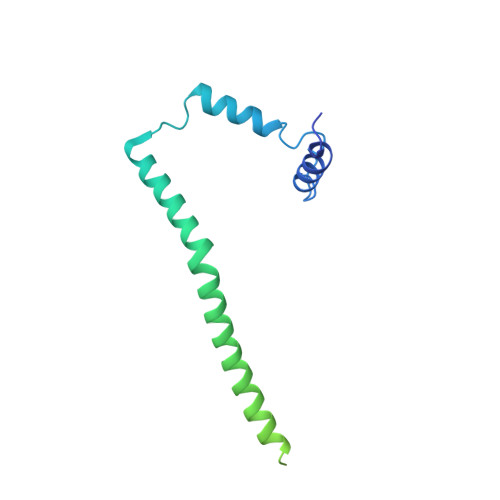
Atomic model for the dimeric FO region of mitochondrial ATP synthase.
Guo, H., Bueler, S.A., Rubinstein, J.L.(2017) Science 358: 936-940
- PubMed: 29074581
- DOI: https://doi.org/10.1126/science.aao4815
- Primary Citation of Related Structures:
6B2Z, 6B8H - PubMed Abstract:
Mitochondrial adenosine triphosphate (ATP) synthase produces the majority of ATP in eukaryotic cells, and its dimerization is necessary to create the inner membrane folds, or cristae, characteristic of mitochondria. Proton translocation through the membrane-embedded F O region turns the rotor that drives ATP synthesis in the soluble F 1 region. Although crystal structures of the F 1 region have illustrated how this rotation leads to ATP synthesis, understanding how proton translocation produces the rotation has been impeded by the lack of an experimental atomic model for the F O region. Using cryo-electron microscopy, we determined the structure of the dimeric F O complex from Saccharomyces cerevisiae at a resolution of 3.6 angstroms. The structure clarifies how the protons travel through the complex, how the complex dimerizes, and how the dimers bend the membrane to produce cristae.
- Hospital for Sick Children Research Institute, Toronto, Ontario M5G 0A4, Canada.
Organizational Affiliation: