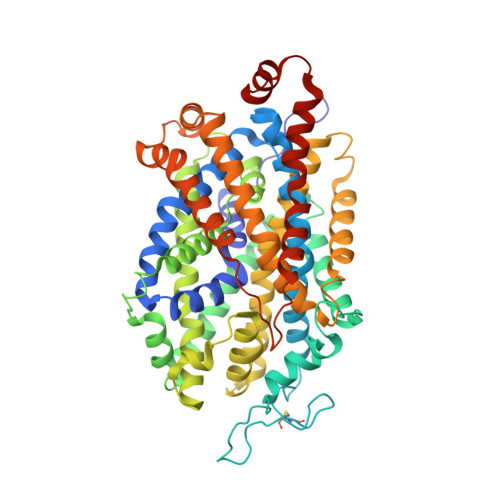
Structural basis for recognition of diverse antidepressants by the human serotonin transporter.
Coleman, J.A., Gouaux, E.(2018) Nat Struct Mol Biol 25: 170-175
- PubMed: 29379174
- DOI: https://doi.org/10.1038/s41594-018-0026-8
- Primary Citation of Related Structures:
6AWN, 6AWO, 6AWP, 6AWQ - PubMed Abstract:
Selective serotonin reuptake inhibitors are clinically prescribed antidepressants that act by increasing the local concentrations of neurotransmitters at synapses and in extracellular spaces via blockade of the serotonin transporter. Here we report X-ray structures of engineered thermostable variants of the human serotonin transporter bound to the antidepressants sertraline, fluvoxamine, and paroxetine. The drugs prevent serotonin binding by occupying the central substrate-binding site and stabilizing the transporter in an outward-open conformation. These structures explain how residues within the central site orchestrate binding of chemically diverse inhibitors and mediate transporter drug selectivity.
- Vollum Institute, Oregon Health & Science University, Portland, OR, USA.
Organizational Affiliation: