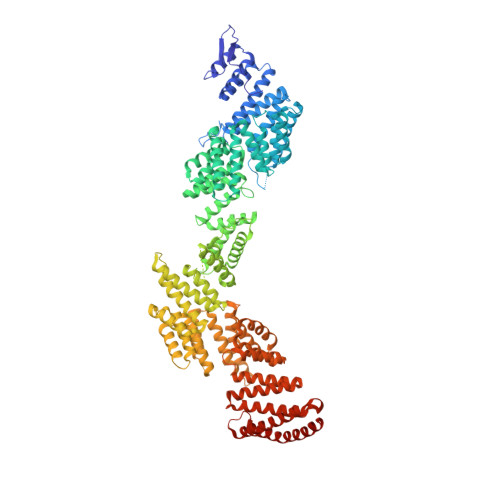
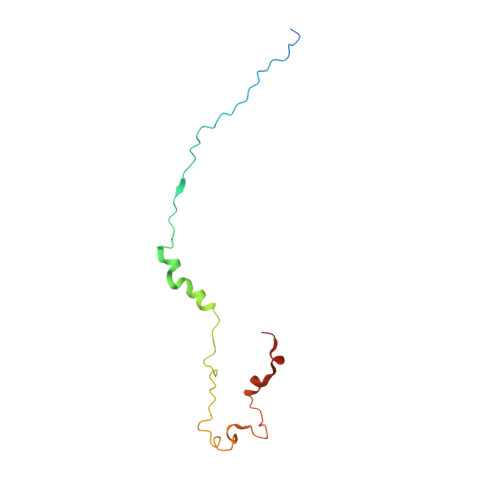
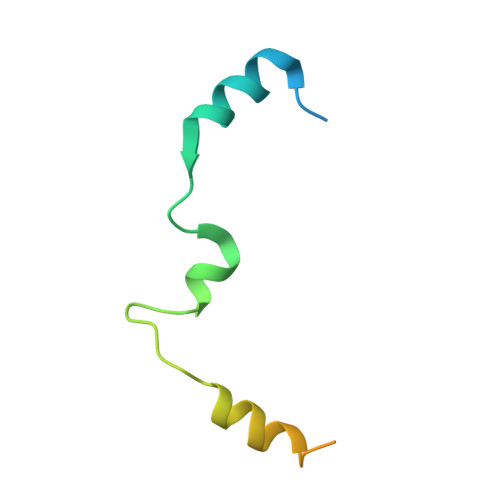
Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Deng, P., Zhou, Y., Jiang, J., Li, H., Tian, W., Cao, Y., Qin, Y., Kim, J., Roeder, R.G., Patel, D.J., Wang, Z.(2018) Proc Natl Acad Sci U S A 115: 9998-10003
- PubMed: 30224485
- DOI: https://doi.org/10.1073/pnas.1812256115
- Primary Citation of Related Structures:
6AF0 - PubMed Abstract:
The polymerase-associated factor 1 (Paf1) complex is a general transcription elongation factor of RNA polymerase II, which is composed of five core subunits, Paf1, Ctr9, Cdc73, Leo1, and Rtf1, and functions as a diverse platform that broadly affects gene expression genome-wide. In this study, we solved the 2.9-Å crystal structure of the core region composed of the Ctr9-Paf1-Cdc73 ternary complex from a thermophilic fungi, which provides a structural perspective of the molecular details of the organization and interactions involving the Paf1 subunits in the core complex. We find that Ctr9 is composed of 21 tetratricopeptide repeat (TPR) motifs that wrap three circular turns in a right-handed superhelical manner around the N-terminal region of an elongated single-polypeptide-chain scaffold of Paf1. The Cdc73 fragment is positioned within the surface groove of Ctr9, where it contacts mainly with Ctr9 and minimally with Paf1. We also identified that the Paf1 complex preferentially binds single-strand-containing DNAs. Our work provides structural insights into the overall architecture of the Paf1 complex and paves the road forward for understanding the molecular mechanisms of the Paf1 complex in transcriptional regulation.
- Key Laboratory of Cell Proliferation and Regulation Biology of Ministry of Education, College of Life Sciences, Beijing Normal University, 100875 Beijing, China.
Organizational Affiliation: