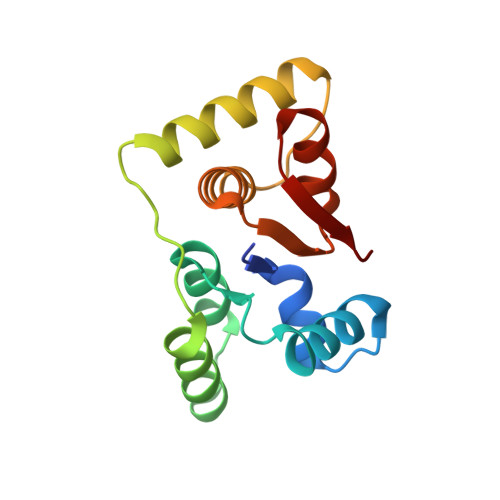
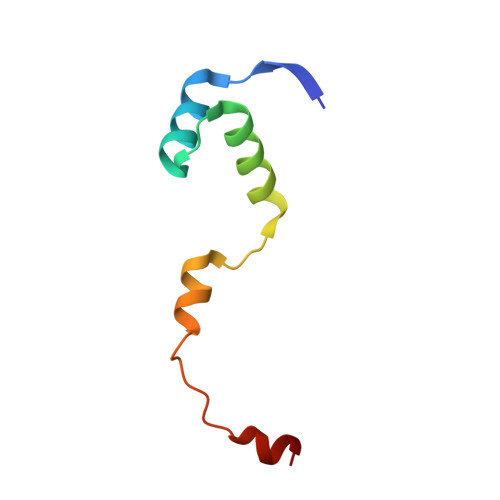
Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: targeting a tRNase to tackle mycobacterial adaptation.
Deep, A., Tiwari, P., Agarwal, S., Kaundal, S., Kidwai, S., Singh, R., Thakur, K.G.(2018) Nucleic Acids Res 46: 11639-11655
- PubMed: 30329074
- DOI: https://doi.org/10.1093/nar/gky924
- Primary Citation of Related Structures:
6A7V - PubMed Abstract:
Toxin-antitoxin (TA) systems are involved in diverse physiological processes in prokaryotes, but their exact role in Mycobacterium tuberculosis (Mtb) virulence and in vivo stress adaptation has not been extensively studied. Here, we demonstrate that the VapBC11 TA module is essential for Mtb to establish infection in guinea pigs. RNA-sequencing revealed that overexpression of VapC11 toxin results in metabolic slowdown, suggesting that modulation of the growth rate is an essential strategy for in vivo survival. Interestingly, overexpression of VapC11 resulted in the upregulation of chromosomal TA genes, suggesting the existence of highly coordinated crosstalk among TA systems. In this study, we also present the crystal structure of the VapBC11 heterooctameric complex at 1.67 Å resolution. Binding kinetic studies suggest that the binding affinities of toxin-substrate and toxin-antitoxin interactions are comparable. We used a combination of structural studies, molecular docking, mutational analysis and in vitro ribonuclease assays to enhance our understanding of the mode of substrate recognition by the VapC11 toxin. Furthermore, we have also designed peptide-based inhibitors to target VapC11 ribonuclease activity. Taken together, we propose that the structure-guided design of inhibitors against in vivo essential ribonucleases might be a novel strategy to hasten clearance of intracellular Mtb.
- Structural Biology Laboratory, G. N. Ramachandran Protein Centre, Council of Scientific and Industrial Research-Institute of Microbial Technology (CSIR-IMTECH), Chandigarh 160036, India.
Organizational Affiliation: