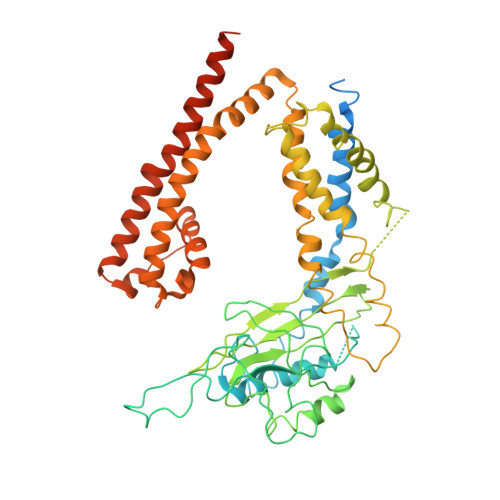
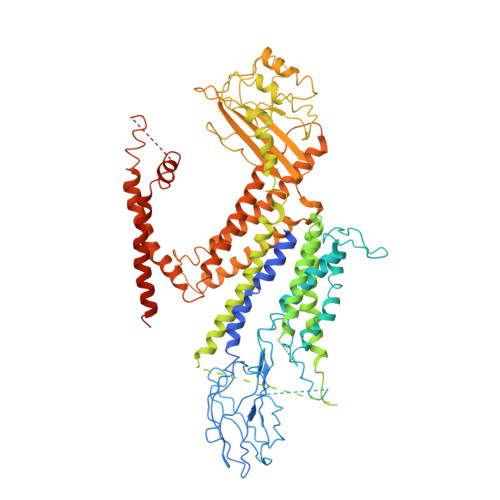
Structure of the human PKD1-PKD2 complex.
Su, Q., Hu, F., Ge, X., Lei, J., Yu, S., Wang, T., Zhou, Q., Mei, C., Shi, Y.(2018) Science 361
- PubMed: 30093605
- DOI: https://doi.org/10.1126/science.aat9819
- Primary Citation of Related Structures:
6A70 - PubMed Abstract:
Mutations in two genes, PKD1 and PKD2 , account for most cases of autosomal dominant polycystic kidney disease, one of the most common monogenetic disorders. Here we report the 3.6-angstrom cryo-electron microscopy structure of truncated human PKD1-PKD2 complex assembled in a 1:3 ratio. PKD1 contains a voltage-gated ion channel (VGIC) fold that interacts with PKD2 to form the domain-swapped, yet noncanonical, transient receptor potential (TRP) channel architecture. The S6 helix in PKD1 is broken in the middle, with the extracellular half, S6a, resembling pore helix 1 in a typical TRP channel. Three positively charged, cavity-facing residues on S6b may block cation permeation. In addition to the VGIC, a five-transmembrane helix domain and a cytosolic PLAT domain were resolved in PKD1. The PKD1-PKD2 complex structure establishes a framework for dissecting the function and disease mechanisms of the PKD proteins.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: