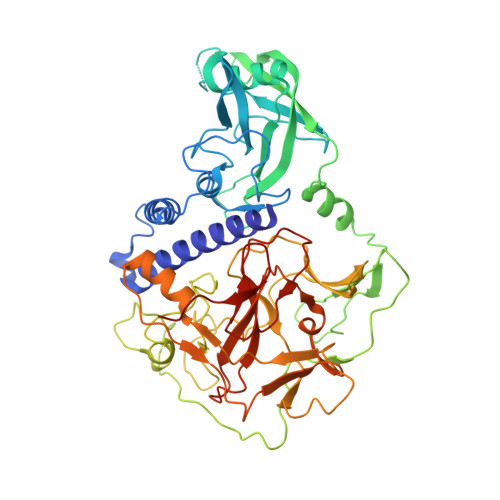
Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Li, X., Harris, C.J., Zhong, Z., Chen, W., Liu, R., Jia, B., Wang, Z., Li, S., Jacobsen, S.E., Du, J.(2018) Proc Natl Acad Sci U S A 115: E8793-E8802
- PubMed: 30150382
- DOI: https://doi.org/10.1073/pnas.1809841115
- Primary Citation of Related Structures:
6A5K, 6A5M, 6A5N - PubMed Abstract:
DNA methylation functions in gene silencing and the maintenance of genome integrity. In plants, non-CG DNA methylation is linked through a self-reinforcing loop with histone 3 lysine 9 dimethylation (H3K9me2). The plant-specific SUPPRESSOR OF VARIEGATION 3-9 HOMOLOG (SUVH) family H3K9 methyltransferases (MTases) bind to DNA methylation marks and catalyze H3K9 methylation. Here, we analyzed the structure and function of Arabidopsis thaliana SUVH6 to understand how this class of enzyme maintains methylation patterns in the genome. We reveal that SUVH6 has a distinct 5-methyl-dC (5mC) base-flipping mechanism involving a thumb loop element. Autoinhibition of H3 substrate entry is regulated by a SET domain loop, and a conformational transition in the post-SET domain upon cofactor binding may control catalysis. In vitro DNA binding and in vivo ChIP-seq data reveal that the different SUVH family H3K9 MTases have distinct DNA binding preferences, targeting H3K9 methylation to sites with different methylated DNA sequences, explaining the context biased non-CG DNA methylation in plants.
- National Key Laboratory of Plant Molecular Genetics, Chinese Academy of Sciences Center for Excellence in Molecular Plant Sciences, Shanghai Center for Plant Stress Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 201602 Shanghai, China.
Organizational Affiliation: