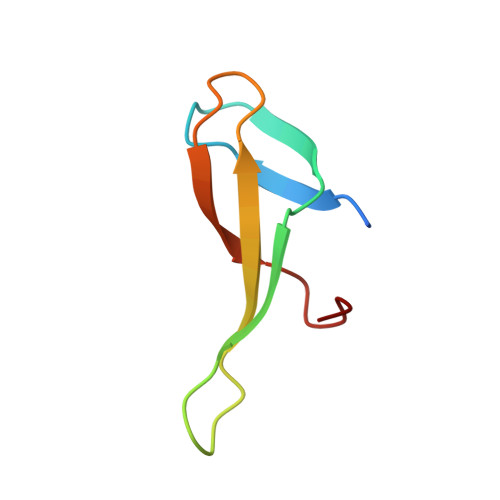
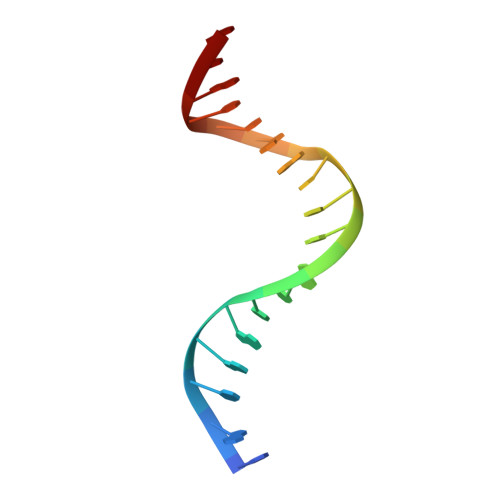
Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Zhang, Z., Zhao, M., Chen, Y., Wang, L., Liu, Q., Dong, Y., Gong, Y., Huang, L.(2019) Mol Microbiol 111: 556-569
- PubMed: 30499242
- DOI: https://doi.org/10.1111/mmi.14173
- Primary Citation of Related Structures:
6A2H, 6A2I - PubMed Abstract:
Archaea have evolved various strategies in chromosomal organization. While histone homologues exist in most archaeal phyla, Cren7 is a chromatin protein conserved in the Crenarchaeota. Here, we show that Cren7 preferentially binds DNA with AT-rich sequences over that with GC-rich sequences with a binding size of 6~7 bp. Structural studies of Cren7 in complex with either an 18-bp or a 20-bp double-stranded DNA fragment reveal that Cren7 binds to the minor groove of DNA as monomers in a head-to-tail manner. The neighboring Cren7 monomers are located on the opposite sides of the DNA duplex, with each introducing a single-step sharp kink by intercalation of the hydrophobic side chain of Leu28, bending the DNA into an S-shape conformation. A structural model for the chromatin fiber folded by Cren7 was established and verified by the analysis of cross-linked Cren7-DNA complexes by atomic force microscopy. Our results suggest that Cren7 differs significantly from Sul7, another chromatin protein conserved among Sulfolobus species, in both DNA binding and deformation. These data shed significant light on the strategy of chromosomal DNA organization in crenarchaea.
- State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, No. 1 West Beichen Road, Chaoyang District, Beijing, 100101, China.
Organizational Affiliation: