Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
Malwal, S.R., Chen, L., Hicks, H., Qu, F., Liu, W., Shillo, A., Law, W.X., Zhang, J., Chandnani, N., Han, X., Zheng, Y., Chen, C.C., Guo, R.T., AbdelKhalek, A., Seleem, M.N., Oldfield, E.(2019) J Med Chem 62: 2564-2581
- PubMed: 30730737
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01878
- Primary Citation of Related Structures:
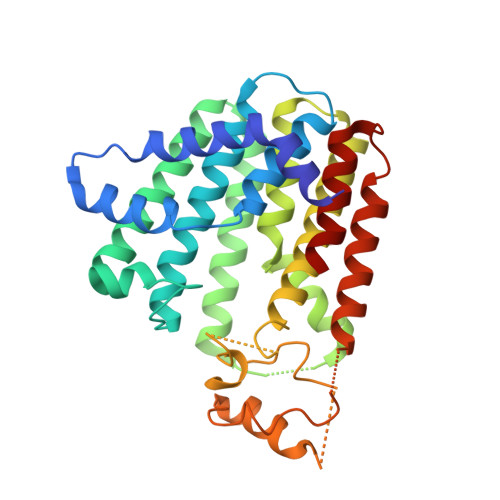
5ZE6, 5ZHE, 5ZLF - PubMed Abstract:
We report that alkyl-substituted bisphosphonates have activity against Bacillus anthracis Sterne (0.40 μg/mL), Mycobacterium smegmatis (1.4 μg/mL), Bacillus subtilis (1.0 μg/mL), and Staphylococcus aureus (13 μg/mL). In many cases, there is no effect of serum binding, as well as low activity against a human embryonic kidney cell line. Targeting of isoprenoid biosynthesis is involved with 74 having IC 50 values of ∼100 nM against heptaprenyl diphosphate synthase and 200 nM against farnesyl diphosphate synthase. B. subtilis growth inhibition was rescued by addition of farnesyl diphosphate, menaquinone-4 (MK-4), or undecaprenyl phosphate (UP), and the combination of MK-4 and UP resulted in a 25× increase in ED 50 , indicating targeting of both quinone and cell wall biosynthesis. Clostridioides difficile was inhibited by 74, and since this organism does not synthesize quinones, cell wall biosynthesis is the likely target. We also solved three X-ray structures of inhibitors bound to octaprenyl diphosphate and/or undecaprenyl diphosphate synthases.
- Industrial Enzymes National Engineering Laboratory , Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences , Tianjin 200208 , China.
Organizational Affiliation: