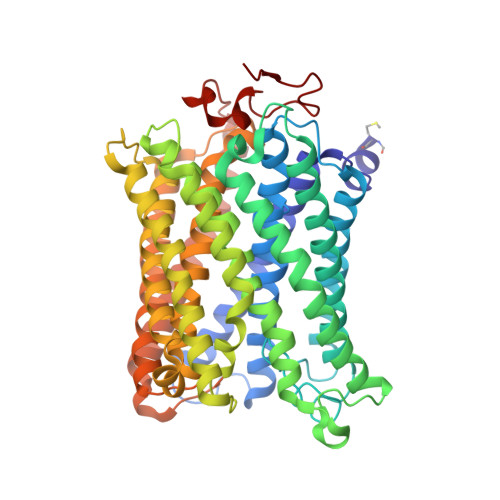
X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
Shimada, A., Hatano, K., Tadehara, H., Yano, N., Shinzawa-Itoh, K., Yamashita, E., Muramoto, K., Tsukihara, T., Yoshikawa, S.(2018) J Biological Chem 293: 14868-14879
- PubMed: 30077971
- DOI: https://doi.org/10.1074/jbc.RA118.003123
- Primary Citation of Related Structures:
5Z84, 5Z85, 5Z86, 5ZCO, 5ZCP, 5ZCQ, 8GVM - PubMed Abstract:
Cytochrome c oxidase (CcO) is the terminal oxidase of cellular respiration, reducing O 2 to water and pumping protons. X-ray structural features have suggested that CcO pumps protons via a mechanism involving electrostatic repulsions between pumping protons in the hydrogen-bond network of a proton-conducting pathway (the H-pathway) and net positive charges created upon oxidation of an iron site, heme a (Fe a 2+ ), for reduction of O 2 at another iron site, heme a 3 (Fe a 3 2+ ). The protons for pumping are transferred to the hydrogen-bond network from the N-side via the water channel of the H-pathway. Back-leakage of protons to the N-side is thought to be blocked by closure of the water channel. To experimentally test this, we examined X-ray structures of the azide-bound, oxidized bovine CcO and found that an azide derivative (N 3 - -Fe a 3 3+ , Cu B 2+ -N 3 - ) induces a translational movement of the heme a 3 plane. This was accompanied by opening of the water channel, revealing that Fe a 3 and the H-pathway are tightly coupled. The channel opening in the oxidized state is likely to induce back-leakage of pumping protons, which lowers the proton level in the hydrogen-bond network during enzymatic turnover. The proton level decrease weakens the electron affinity of Fe a , if Fe a electrostatically interacts with protons in the hydrogen-bond network. The previously reported azide-induced redox-potential decrease in Fe a supports existence of the electrostatic interaction. In summary, our results indicate that the H-pathway is critical for CcO's proton-pumping function.
- From the Picobiology Institute and.
Organizational Affiliation: