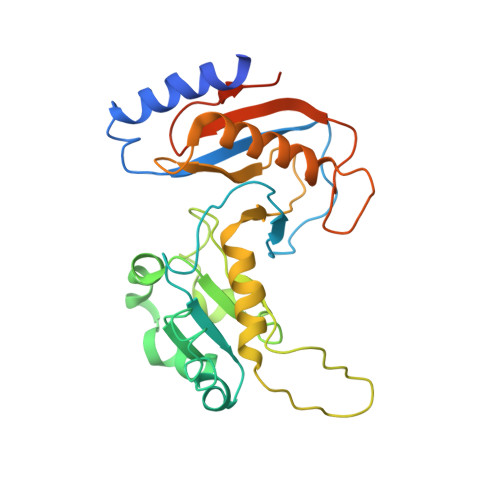
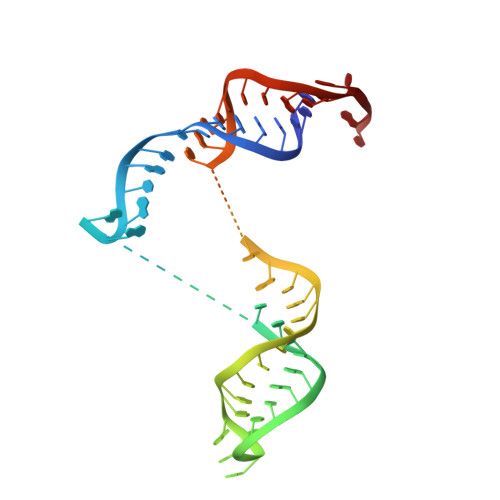
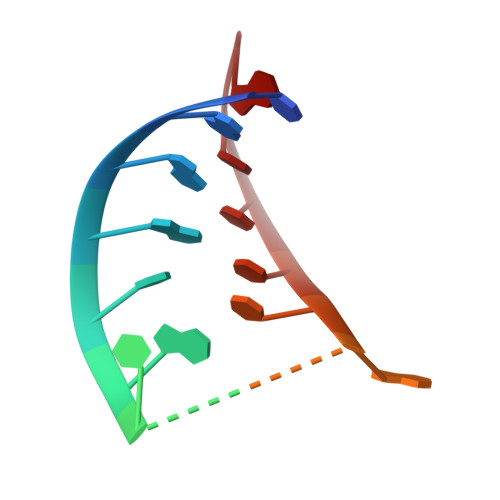
Structure and RNA recognition of ribosome assembly factor Utp30.
Hu, J., Zhu, X., Ye, K.(2017) RNA 23: 1936-1945
- PubMed: 28951391
- DOI: https://doi.org/10.1261/rna.062695.117
- Primary Citation of Related Structures:
5YDT, 5YDU - PubMed Abstract:
The 90S preribosomes are gigantic early assembly intermediates of small ribosomal subunits. Cryo-EM structures of 90S were recently determined, but many of its components have not been accurately modeled. Here we determine the crystal structure of yeast Utp30, a ribosomal L1 domain-containing protein in 90S, at 2.65 Å resolution, revealing a classic two-domain fold. The structure of Utp30 fits well into the cryo-EM density of 90S, confirming its previously assigned location. Utp30 binds to the rearranged helix 41 of 18S rRNA and helix 4 of 5' external transcribed spacer in 90S. Comparison of RNA-binding modes of different L1 domains illustrates that they consistently recognize a short RNA duplex with the concaved surface of domain I, but are versatile in RNA recognition outside the core interface. Cic1 is a paralog of Utp30 associating with large subunit preribosomes. Utp30 and Cic1 share similar RNA-binding modes, suggesting that their distinct functions may be executed by a single protein in other organisms. Deletion of Utp30 does not affect the composition of 90S. The nonessential role of Utp30 could be ascribed to its peripheral localization and redundant interactions in 90S.
- Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: