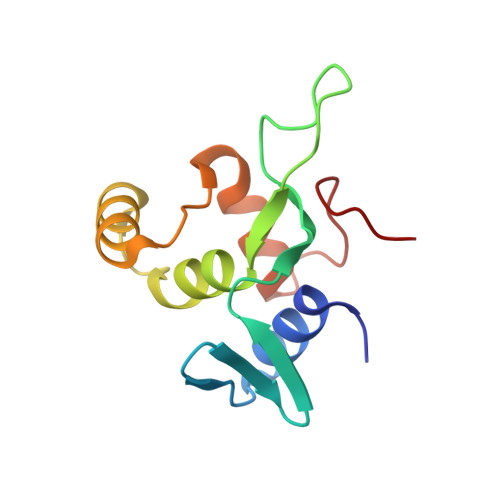
Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sun, W., Kuang, X.L., Liu, Y.P., Tian, L.F., Yan, X.X., Xu, W.(2017) Sci Rep 7: 15638-15638
- PubMed: 29142233
- DOI: https://doi.org/10.1038/s41598-017-15715-9
- Primary Citation of Related Structures:
5YDE, 5YDF - PubMed Abstract:
CDC73/Parafibromin is a critical component of the Paf1 complex (PAF1C), which is involved in transcriptional elongation and histone modifications. Mutations of the human CDC73/HRPT2 gene are associated with hyperparathyroidism-jaw tumor (HPT-JT) syndrome, an autosomal dominant disorder. CDC73/parafibromin was initially recognized as a tumor suppressor by inhibiting cell proliferation via repression of cyclin D1 and c-myc genes. In recent years, it has also shown oncogenic features by activating the canonical Wnt/β-catenin signal pathway. Here, through limited proteolysis analysis, we demonstrate that the evolutionarily conserved human CDC73 N-terminal 111 residues form a globularly folded domain (hCDC73-NTD). We have determined a crystal structure of hCDC73-NTD at 1.02 Å resolution, which reveals a novel protein fold. CDC73-NTD contains an extended hydrophobic groove on its surface that may be important for its function. Most pathogenic CDC73 missense mutations associated with the HPT-JT syndrome are located in the region encoding CDC73-NTD. Our crystal and biochemical data indicate that most CDC73 missense mutations disrupt the folding of the hydrophobic core of hCDC73-NTD, while others such as the K34Q mutant reduce its thermostability. Overall, our results provide a solid structural basis for understanding the structure and function of CDC73 and its association with the HPT-JT syndrome and other diseases.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, P. R. China.
Organizational Affiliation: