Molecular basis for unique specificity of human TRAF4 for platelets GPIb beta and GPVI
Kim, C.M., Son, Y.J., Kim, S., Kim, S.Y., Park, H.H.(2017) Proc Natl Acad Sci U S A 114: 11422-11427
- PubMed: 29073066
- DOI: https://doi.org/10.1073/pnas.1708688114
- Primary Citation of Related Structures:
5YC1 - PubMed Abstract:
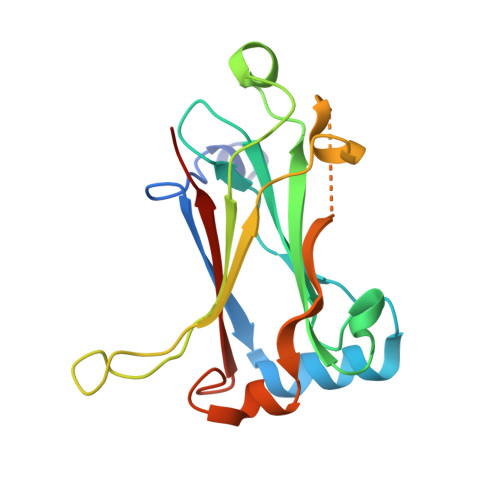
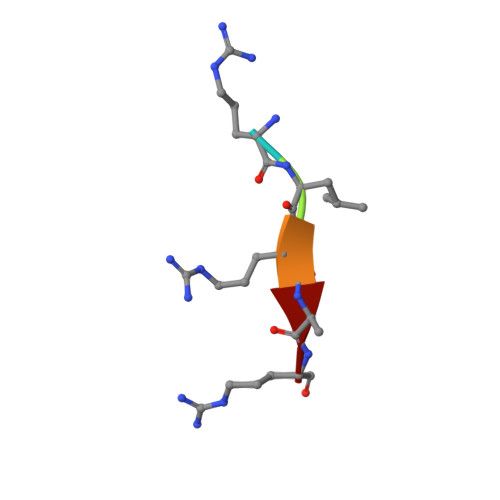
Tumor necrosis factor (TNF)-receptor associated factor 4 (TRAF4), an adaptor protein with E3-ligase activity, is involved in embryogenesis, cancer initiation and progression, and platelet receptor (GPIb-IX-V complex and GPVI)-mediated signaling for reactive oxygen species (ROS) production that initiates thrombosis at arterial shears. Disruption of platelet receptors and the TRAF4 interaction is a potential target for therapeutic intervention by antithrombotic drugs. Here, we report a crystal structure of TRAF4 (amino acid residues 290∼470) in complex with a peptide from the GPIbβ receptor (amino acid residues 177∼181). The GPIbβ peptide binds to a unique shallow surface composed of two hydrophobic pockets on TRAF4. Further studies revealed the TRAF4-binding motif Arg-Leu-X-Ala. The TRAF4-binding motif was present not only in platelet receptors but also in the TGF-β receptor. The current structure will provide a template for furthering our understanding of the receptor-binding specificity of TRAF4, TRAF4-mediated signaling, and related diseases.
- School of Natural Science, Department of Chemistry and Biochemistry, Yeungnam University, Gyeongsan 712-749, South Korea.
Organizational Affiliation: