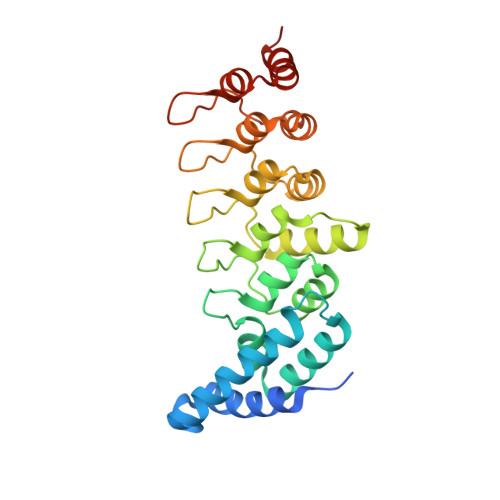
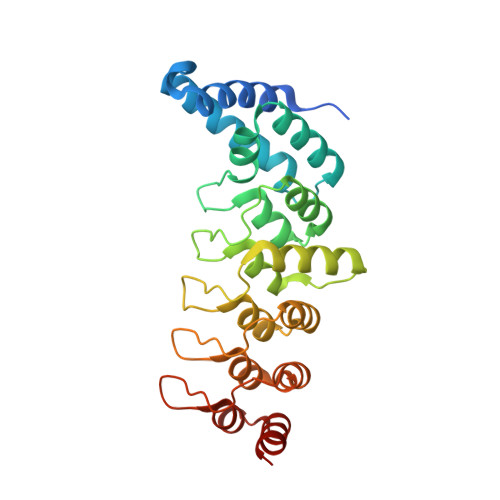
Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
Guo, Q., Liao, S., Zhu, Z., Li, Y., Li, F., Xu, C.(2018) J Biological Chem 293: 557-566
- PubMed: 29183992
- DOI: https://doi.org/10.1074/jbc.M117.817494
- Primary Citation of Related Structures:
5YBJ, 5YBU, 5YBV - PubMed Abstract:
A well-controlled microtubule organization is essential for intracellular transport, cytoskeleton maintenance, and cell development. KN motif and ankyrin repeat domain-containing protein 1 (KANK1), a member of KANK family, recruits kinesin family member 21A (KIF21A) to the cell cortex to control microtubule growth via its C-terminal ankyrin domain. However, how the KANK1 ankyrin domain recognizes KIF21A and whether other KANK proteins can also bind KIF21A remain unknown. Here, using a combination of structural, site-directed mutagenesis, and biochemical studies, we found that a stretch of ∼22 amino acids in KIF21A is sufficient for binding to KANK1 and its close homolog KANK2. We further solved the complex structure of the KIF21A peptide with either the KANK1 ankyrin domain or the KANK2 ankyrin domain. In each complex, KIF21A is recognized by two distinct pockets of the ankyrin domain and adopts helical conformations upon binding to the ankyrin domain. The elucidated KANK structures may advance our understanding of the role of KANK1 as a scaffolding molecule in controlling microtubule growth at the cell periphery.
- From the Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei 230027, China and.
Organizational Affiliation: