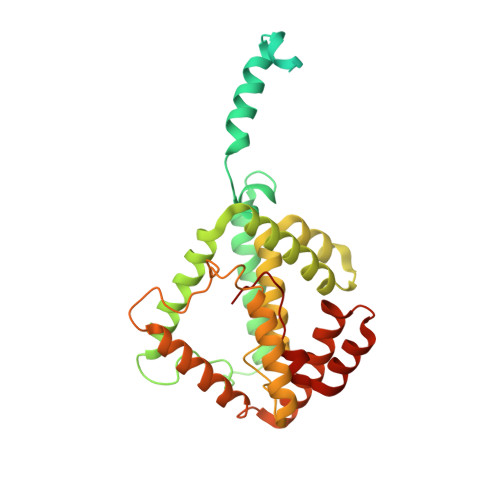
PUB Query on PUB Download Ideal Coordinates CCD File ABB [auth t5]
ABB [auth t5],
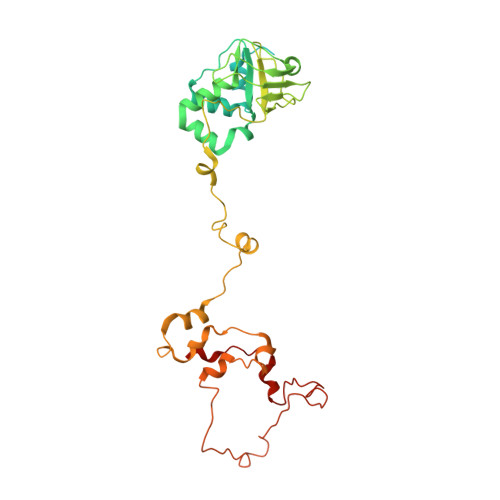
PHYCOUROBILIN 33 H42 N4 O6 PEB Query on PEB Download Ideal Coordinates CCD File AAB [auth h5]
AAB [auth h5],
PHYCOERYTHROBILIN 33 H40 N4 O6