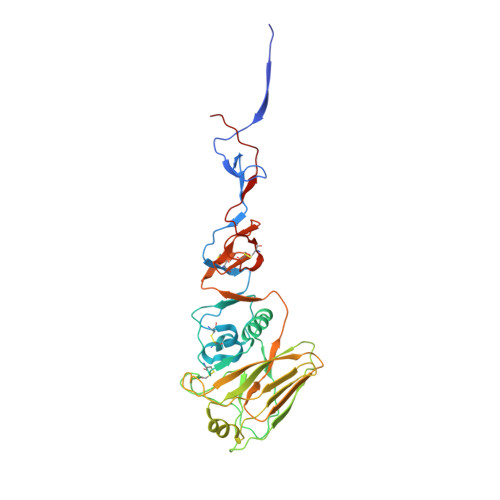
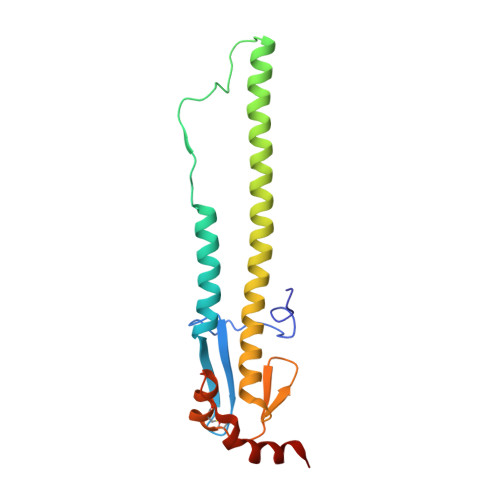
A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA.
Bangaru, S., Zhang, H., Gilchuk, I.M., Voss, T.G., Irving, R.P., Gilchuk, P., Matta, P., Zhu, X., Lang, S., Nieusma, T., Richt, J.A., Albrecht, R.A., Vanderven, H.A., Bombardi, R., Kent, S.J., Ward, A.B., Wilson, I.A., Crowe, J.E.(2018) Nat Commun 9: 2669-2669
- PubMed: 29991715
- DOI: https://doi.org/10.1038/s41467-018-04704-9
- Primary Citation of Related Structures:
5W42, 5XRQ, 5XRS, 5XRT - PubMed Abstract:
The high rate of antigenic drift in seasonal influenza viruses necessitates frequent changes in vaccine composition. Recent seasonal H3 vaccines do not protect against swine-origin H3N2 variant (H3N2v) strains that recently have caused severe human infections. Here, we report a human V H 1-69 gene-encoded monoclonal antibody (mAb) designated H3v-47 that exhibits potent cross-reactive neutralization activity against human and swine H3N2 viruses that circulated since 1989. The crystal structure and electron microscopy reconstruction of H3v-47 Fab with the H3N2v hemagglutinin (HA) identify a unique epitope spanning the vestigial esterase and receptor-binding subdomains that is distinct from that of any known neutralizing antibody for influenza A H3 viruses. MAb H3v-47 functions largely by blocking viral egress from infected cells. Interestingly, H3v-47 also engages Fcγ receptor and mediates antibody dependent cellular cytotoxicity (ADCC). This newly identified conserved epitope can be used in design of novel immunogens for development of broadly protective H3 vaccines.
- Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN, 37232, USA.
Organizational Affiliation: