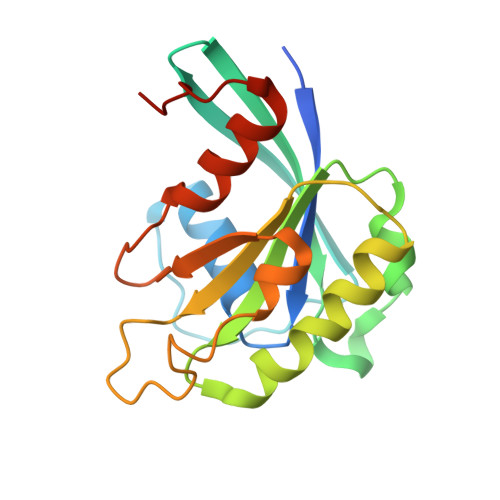
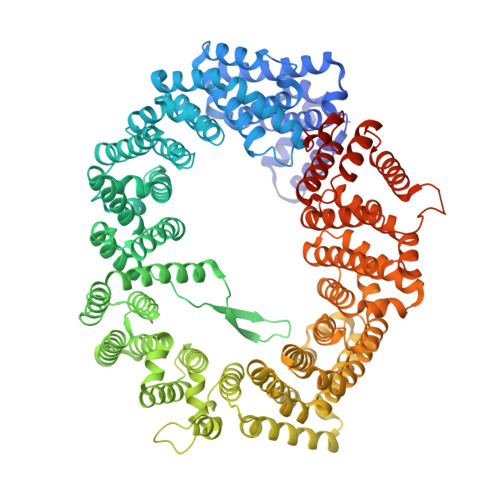
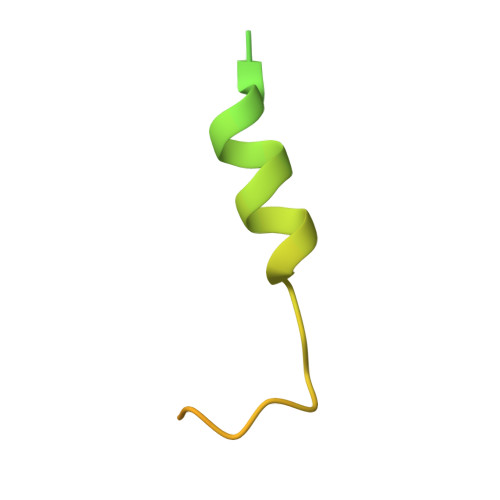
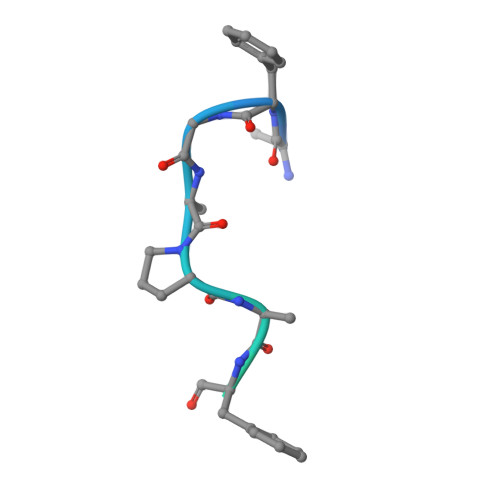
Crystal structure of the Xpo1p nuclear export complex bound to the SxFG/PxFG repeats of the nucleoporin Nup42p
Koyama, M., Hirano, H., Shirai, N., Matsuura, Y.(2017) Genes Cells 22: 861-875
- PubMed: 28791779
- DOI: https://doi.org/10.1111/gtc.12520
- Primary Citation of Related Structures:
5XOJ - PubMed Abstract:
Xpo1p (yeast CRM1) is the major nuclear export receptor that carries a plethora of proteins and ribonucleoproteins from the nucleus to cytoplasm. The passage of the Xpo1p nuclear export complex through nuclear pore complexes (NPCs) is facilitated by interactions with nucleoporins (Nups) containing extensive repeats of phenylalanine-glycine (so-called FG repeats), although the precise role of each Nup in the nuclear export reaction remains incompletely understood. Here we report structural and biochemical characterization of the interactions between the Xpo1p nuclear export complex and the FG repeats of Nup42p, a nucleoporin localized at the cytoplasmic face of yeast NPCs and has characteristic SxFG/PxFG sequence repeat motif. The crystal structure of Xpo1p-PKI-Nup42p-Gsp1p-GTP complex identified three binding sites for the SxFG/PxFG repeats on HEAT repeats 14-20 of Xpo1p. Mutational analyses of Nup42p showed that the conserved serines and prolines in the SxFG/PxFG repeats contribute to Xpo1p-Nup42p binding. Our structural and biochemical data suggest that SxFG/PxFG-Nups such as Nup42p and Nup159p at the cytoplasmic face of NPCs provide high-affinity docking sites for the Xpo1p nuclear export complex in the terminal stage of NPC passage and that subsequent disassembly of the nuclear export complex facilitates recycling of free Xpo1p back to the nucleus.
- Division of Biological Science, Nagoya University, Nagoya, 464-8602, Japan.
Organizational Affiliation: