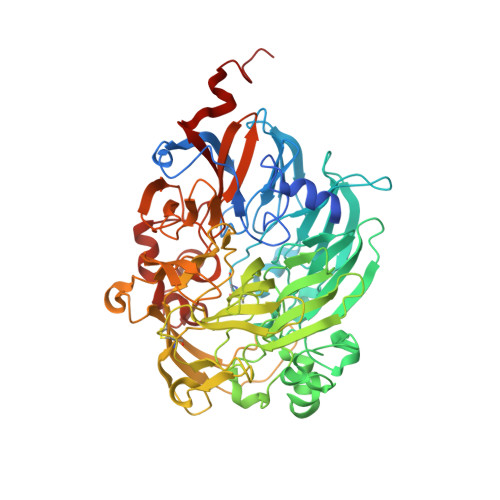
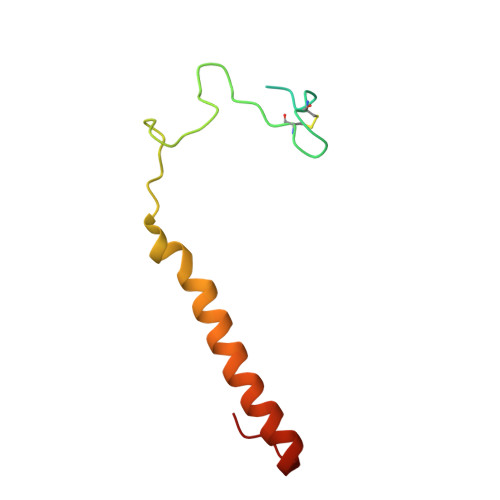
The crystal structure of methanol dehydrogenase, a quinoprotein from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT
Cao, T.P., Choi, J.M., Kim, S.W., Lee, S.H.(2018) J Microbiol 56: 246-254
- PubMed: 29492864
- DOI: https://doi.org/10.1007/s12275-018-7483-y
- Primary Citation of Related Structures:
5XM3 - PubMed Abstract:
The first crystal structure of a pyrroloquinoline quinone (PQQ)-dependent methanol dehydrogenase (MDH) from a marine methylotrophic bacterium, Methylophaga aminisulfidivorans MP T (MDH Mas ), was determined at 1.7 Å resolution. The active form of MDH Mas (or MDHI Mas ) is a heterotetrameric α 2 β 2 , where each β-subunit assembles on one side of each of the α-subunits, in a symmetrical fashion, so that two β-subunits surround the two PQQ-binding pockets on the α-subunits. The active site consists of a PQQ molecule surrounded by a β-propeller fold for each α-subunit. Interestingly, the PQQ molecules are coordinated by a Mg 2+ ion, instead of the Ca 2+ ion that is commonly found in the terrestrial MDHI, indicating the efficiency of osmotic balance regulation in the high salt environment. The overall interaction of the β-subunits with the α-subunits appears tighter than that of terrestrial homologues, suggesting the efficient maintenance of MDHI Mas integrity in the sea water environment to provide a firm basis for complex formation with MxaJ Mas or Cyt c L . With the help of the features mentioned above, our research may enable the elucidation of the full molecular mechanism of methanol oxidation by taking advantage of marine bacterium-originated proteins in the methanol oxidizing system (mox), including MxaJ, as the attainment of these proteins from terrestrial bacteria for structural studies has not been successful.
- Department of Cellular and Molecular Medicine, Chosun University School of Medicine, Gwangju, 61452, Republic of Korea.
Organizational Affiliation: