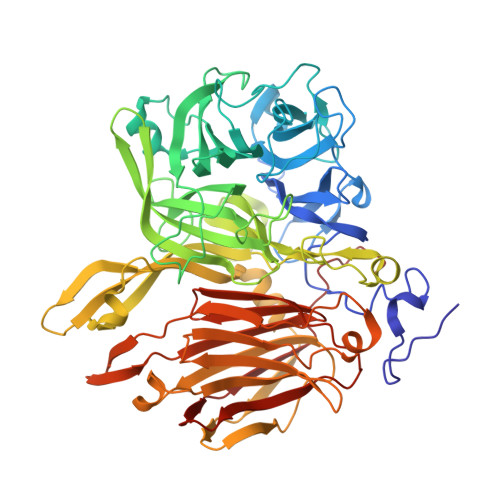
Crystal structure of a beta-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii
Nagaya, M., Kimura, M., Gozu, Y., Sato, S., Hirano, K., Tochio, T., Nishikawa, A., Tonozuka, T.(2017) Biosci Biotechnol Biochem 81: 1786-1795
- PubMed: 28715279
- DOI: https://doi.org/10.1080/09168451.2017.1353405
- Primary Citation of Related Structures:
5XH8, 5XH9, 5XHA - PubMed Abstract:
β-Fructofuranosidases belonging to glycoside hydrolase family (GH) 32 are enzymes that hydrolyze sucrose. Some GH32 enzymes also catalyze transfructosylation to produce fructooligosaccharides. We found that Aspergillus kawachii IFO 4308 β-fructofuranosidase (AkFFase) produces fructooligosaccharides, mainly 1-kestose, from sucrose. We determined the crystal structure of AkFFase. AkFFase is composed of an N-terminal small component, a β-propeller catalytic domain, an α-helical linker, and a C-terminal β-sandwich, similar to other GH32 enzymes. AkFFase forms a dimer, and the dimerization pattern is different from those of other oligomeric GH32 enzymes. The complex structure of AkFFase with fructose unexpectedly showed that fructose binds both subsites -1 and +1, despite the fact that the catalytic residues were not mutated. Fructose at subsite +1 interacts with Ile146 and Glu296 of AkFFase via direct hydrogen bonds.
- a Department of Applied Biological Science , Tokyo University of Agriculture and Technology , Fuchu , Japan.
Organizational Affiliation: