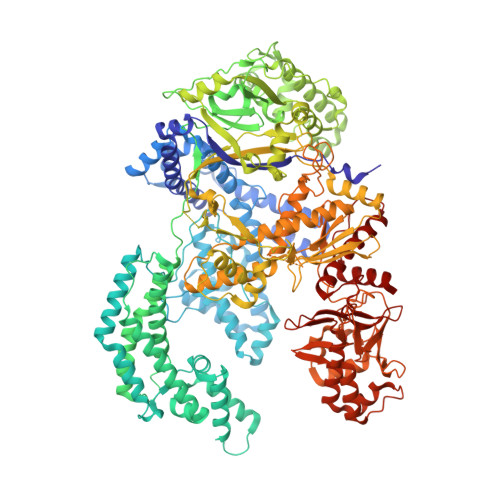
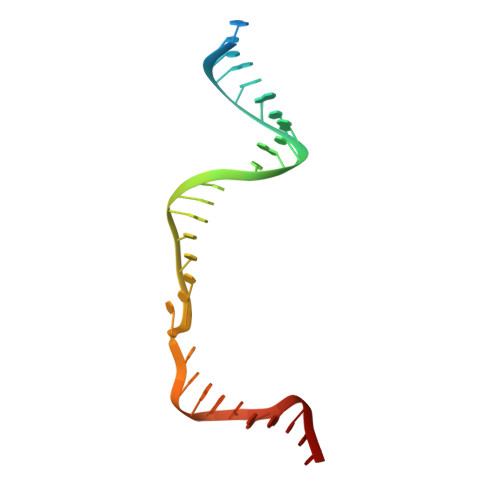
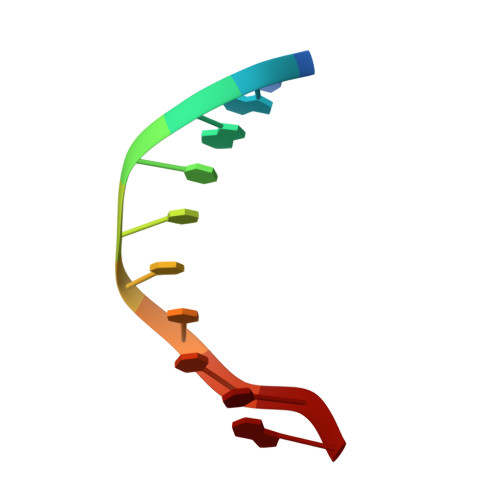
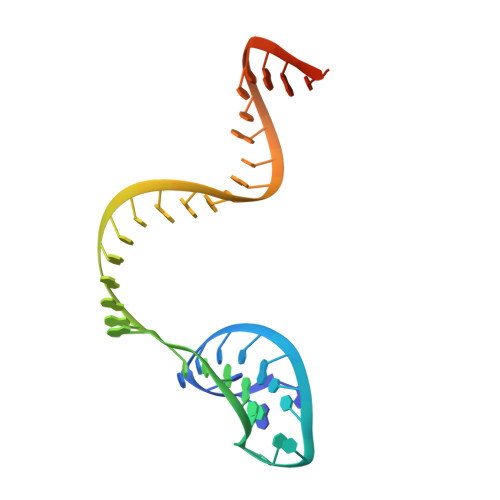Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Nishimasu, H., Yamano, T., Gao, L., Zhang, F., Ishitani, R., Nureki, O.(2017) Mol Cell 67: 139-147.e2
- PubMed: 28595896
- DOI: https://doi.org/10.1016/j.molcel.2017.04.019
- Primary Citation of Related Structures:
5XH6, 5XH7 - PubMed Abstract:
The RNA-guided Cpf1 nuclease cleaves double-stranded DNA targets complementary to the CRISPR RNA (crRNA), and it has been harnessed for genome editing technologies. Recently, Acidaminococcus sp. BV3L6 (AsCpf1) was engineered to recognize altered DNA sequences as the protospacer adjacent motif (PAM), thereby expanding the target range of Cpf1-mediated genome editing. Whereas wild-type AsCpf1 recognizes the TTTV PAM, the RVR (S542R/K548V/N552R) and RR (S542R/K607R) variants can efficiently recognize the TATV and TYCV PAMs, respectively. However, their PAM recognition mechanisms remained unknown. Here we present the 2.0 Å resolution crystal structures of the RVR and RR variants bound to a crRNA and its target DNA. The structures revealed that the RVR and RR variants primarily recognize the PAM-complementary nucleotides via the substituted residues. Our high-resolution structures delineated the altered PAM recognition mechanisms of the AsCpf1 variants, providing a basis for the further engineering of CRISPR-Cpf1.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan; JST, PRESTO, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan. Electronic address: nisimasu@bs.s.u-tokyo.ac.jp.
Organizational Affiliation: