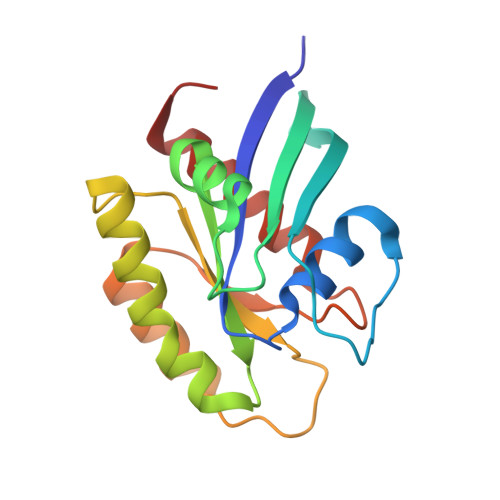
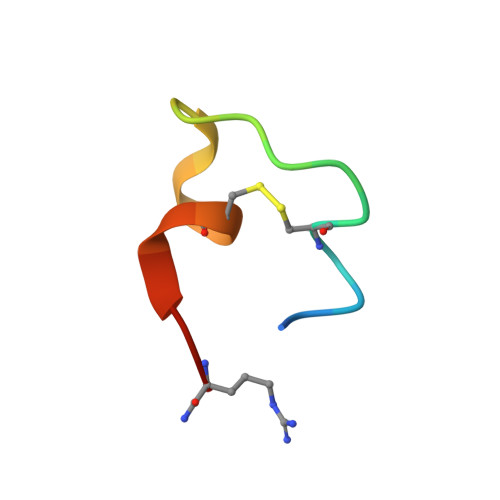
Crystal Structure of a Human K-Ras G12D Mutant in Complex with GDP and the Cyclic Inhibitory Peptide KRpep-2d
Sogabe, S., Kamada, Y., Miwa, M., Niida, A., Sameshima, T., Kamaura, M., Yonemori, K., Sasaki, S., Sakamoto, J., Sakamoto, K.(2017) ACS Med Chem Lett 8: 732-736
- PubMed: 28740607
- DOI: https://doi.org/10.1021/acsmedchemlett.7b00128
- Primary Citation of Related Structures:
5XCO - PubMed Abstract:
The Ras proteins play roles in cell differentiation, proliferation, and survival. Aberrant signaling through Ras-mediated pathways in tumor cells occurs as a result of several types of mutational damage, which most frequently affects the amino acids G12, G13, and Q61. Recently, KRpep-2d was identified as a K-Ras(G12D) selective inhibitory peptide against the G12D mutant of K-Ras, which is a key member of the Ras protein family and an attractive cancer therapeutic target. In this study, the crystal structure of the human K-Ras(G12D) mutant was determined in complex with GDP and KRpep-2d at 1.25 Å resolution. This structure revealed that the peptide binds near Switch II and allosterically blocks protein-protein interactions with the guanine nucleotide exchange factor. This discovery of a unique binding pocket provides valuable information that will facilitate the design of direct Ras inhibitors.
- Pharmaceutical Research Division, Takeda Pharmaceutical Company Limited, 26-1, Muraoka-Higashi 2-chome, Fujisawa, Kanagawa 251-8555, Japan.
Organizational Affiliation: