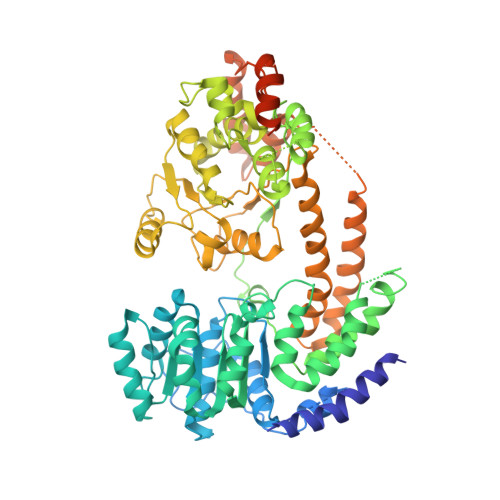
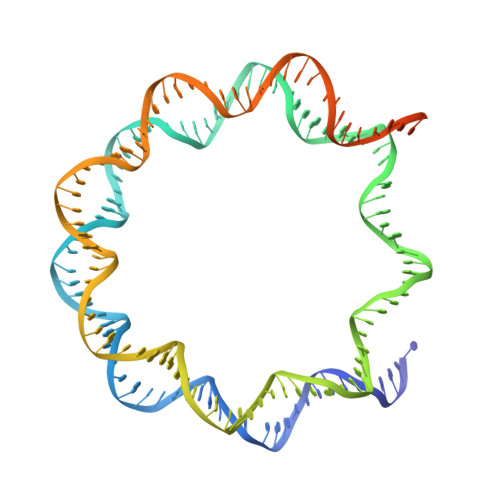
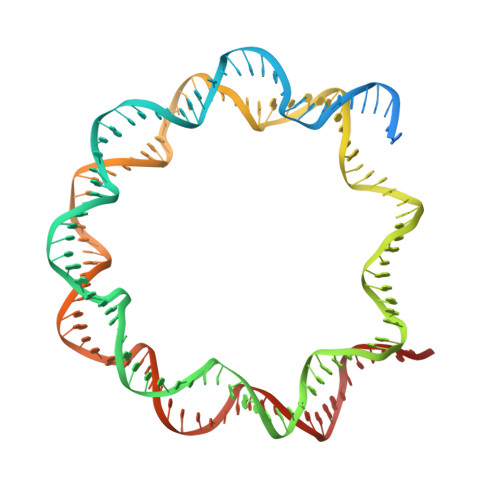
Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Liu, X., Li, M., Xia, X., Li, X., Chen, Z.(2017) Nature 544: 440-445
- PubMed: 28424519
- DOI: https://doi.org/10.1038/nature22036
- Primary Citation of Related Structures:
5X0X, 5X0Y - PubMed Abstract:
Chromatin remodellers are helicase-like, ATP-dependent enzymes that alter chromatin structure and nucleosome positions to allow regulatory proteins access to DNA. Here we report the cryo-electron microscopy structure of chromatin remodeller Switch/sucrose non-fermentable (SWI2/SNF2) from Saccharomyces cerevisiae bound to the nucleosome. The structure shows that the two core domains of Snf2 are realigned upon nucleosome binding, suggesting activation of the enzyme. The core domains contact each other through two induced Brace helices, which are crucial for coupling ATP hydrolysis to chromatin remodelling. Snf2 binds to the phosphate backbones of one DNA gyre of the nucleosome mainly through its helicase motifs within the major domain cleft, suggesting a conserved mechanism of substrate engagement across different remodellers. Snf2 contacts the second DNA gyre via a positively charged surface, providing a mechanism to anchor the remodeller at a fixed position of the nucleosome. Snf2 locally deforms nucleosomal DNA at the site of binding, priming the substrate for the remodelling reaction. Together, these findings provide mechanistic insights into chromatin remodelling.
- Ministry of Education Key Laboratory of Protein Science, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: