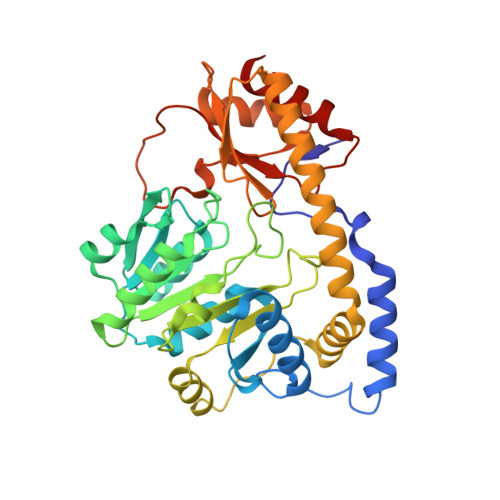
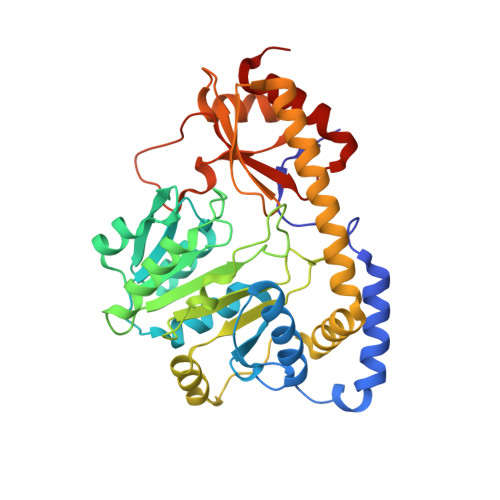
Crystal structure of the C-terminal domain of Bacillus subtilis GabR reveals a closed conformation by gamma-aminobutyric acid binding, inducing transcriptional activation
Park, S.A., Park, Y.S., Lee, K.S.(2017) Biochem Biophys Res Commun 487: 287-291
- PubMed: 28412355
- DOI: https://doi.org/10.1016/j.bbrc.2017.04.052
- Primary Citation of Related Structures:
5X03 - PubMed Abstract:
Bacillus subtilis GabR (BsGabR) is involved in the γ-aminobutyric acid (GABA) catabolism as a transcriptional regulator, consisting of an N-terminal helix-turn-helix DNA-binding domain and a C-terminal aminotransferase-like (AT-like) domain. Research on the C-terminal AT-like domain of BsGabR (BsGabR-CTD) has focused on the interaction with GABA as an effector, but most its functional details remain unclear. To understand the underlying mechanism, we report the crystal structure of BsGabR-CTD in complex with pyridoxal 5'-phosphate (PLP) and GABA at 2.0 Å resolution. The structure of ligand-bound BsGabR-CTD revealed two distinct monomeric states in a homodimer. One subunit is a closed-form containing the PLP-GABA adduct, and the other subunit is a PLP-bound open-form. Our structural studies provide a detailed mechanism indicating that the open-to-closed transition by the binding of GABA induces the conformational rearrangement of BsGabR-CTD, which may trigger the activation of transcription.
- Department of Clinical Laboratory Science, College of Health Sciences, Catholic University of Pusan, Busan 46252, Republic of Korea.
Organizational Affiliation: