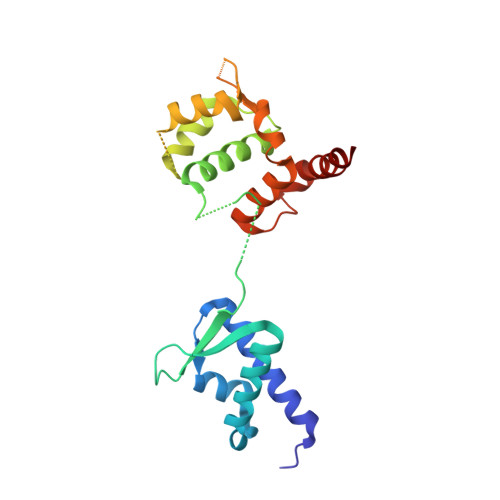
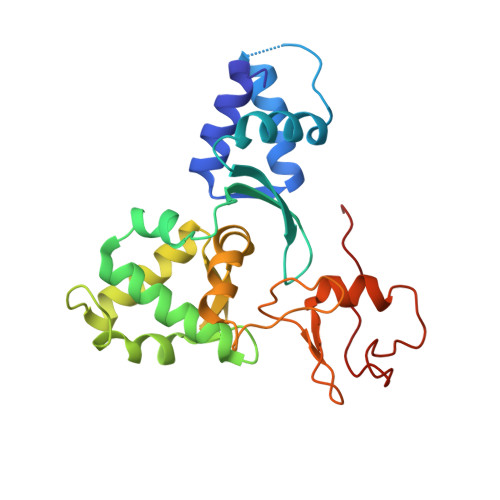
Mage-Ring Protein Complexes Comprise A Family Of E3 Ubiquitin Ligases.
Doyle, J.M., Gao, J., Wang, J., Yang, M., Potts, P.R.(2010) Mol Cell 39: 963-974
- PubMed: 20864041
- DOI: https://doi.org/10.1016/j.molcel.2010.08.029
- Primary Citation of Related Structures:
5WY5 - PubMed Abstract:
The melanoma antigen (MAGE) family consists of more than 60 genes, many of which are cancer-testis antigens that are highly expressed in cancer and play a critical role in tumorigenesis. However, the biochemical and cellular functions of this enigmatic family of proteins have remained elusive. Here, we identify really interesting new gene (RING) domain proteins as binding partners for MAGE family proteins. Multiple MAGE family proteins bind E3 RING ubiquitin ligases with specificity. The crystal structure of one of these MAGE-RING complexes, MAGE-G1-NSE1, reveals structural insights into MAGE family proteins and their interaction with E3 RING ubiquitin ligases. Biochemical and cellular assays demonstrate that MAGE proteins enhance the ubiquitin ligase activity of RING domain proteins. For example, MAGE-C2-TRIM28 is shown to target p53 for degradation in a proteasome-dependent manner, consistent with its tumorigenic functions. These findings define a biochemical and cellular function for the MAGE protein family.
- Department of Biochemistry, University of Texas Southwestern Medical Center, Dallas, TX 75390-9038, USA.
Organizational Affiliation: