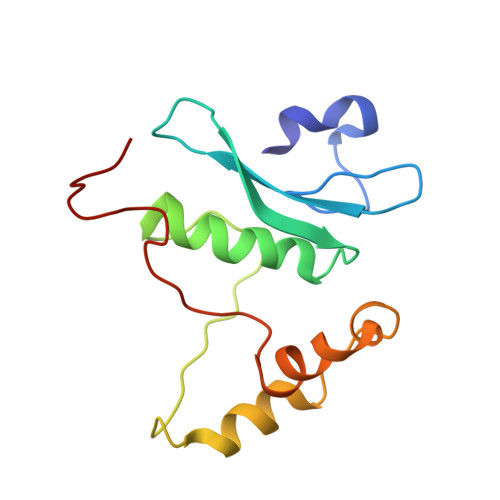
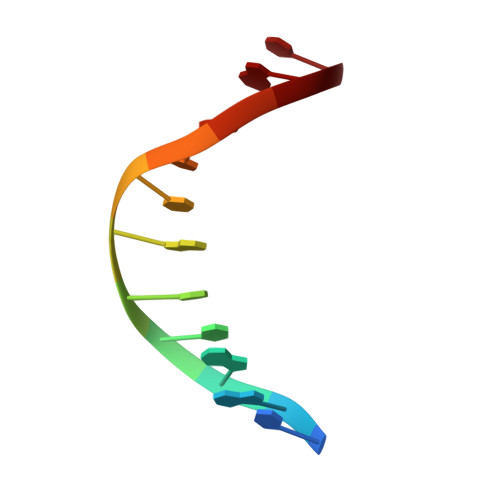
Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box.
Chen, C.Y., Lin, P.H., Chen, K.H., Cheng, Y.S.(2020) Plant Mol Biol 104: 483-498
- PubMed: 32813232
- DOI: https://doi.org/10.1007/s11103-020-01052-5
- Primary Citation of Related Structures:
5WX9 - PubMed Abstract:
The phytohormone ethylene is widely involved in many developmental processes and is a crucial regulator of defense responses against biotic and abiotic stresses in plants. Ethylene-responsive element binding protein, a member of the APETALA2/ethylene response factor (AP2/ERF) superfamily, is a transcription factor that regulates stress-responsive genes by recognizing a specific cis-acting element of target DNA. A previous study showed only the NMR structure of the AP2/ERF domain of AtERF100 in complex with a GCC box DNA motif. In this report, we determined the crystal structure of AtERF96 in complex with a GCC box at atomic resolution. We analyzed the binding residues of the conserved AP2/ERF domain in the DNA recognition sequence. In addition to the AP2/ERF domain, an N-terminal α-helix of AtERF96 participates in DNA interaction in the flanking region. We also demonstrated the structure of AtERF96 EDLL motif, a unique conserved motif in the group IX of AP2/ERF family, might involve in the transactivation of defense-related genes. Our study establishes the structural basis of the AtERF96 transcription factor in complex with the GCC box, as well as the DNA binding mechanisms of the N-terminal α-helix and AP2/ERF domain.
- Institute of Plant Biology, National Taiwan University, Taipei, Taiwan.
Organizational Affiliation: