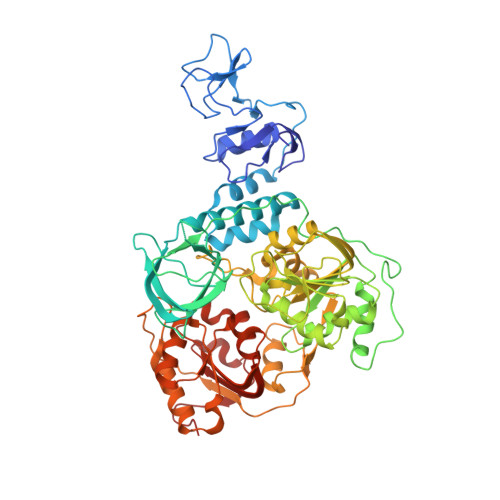
Crystal structure of Middle East respiratory syndrome coronavirus helicase
Hao, W., Wojdyla, J.A., Zhao, R., Han, R., Das, R., Zlatev, I., Manoharan, M., Wang, M., Cui, S.(2017) PLoS Pathog 13: e1006474-e1006474
- PubMed: 28651017
- DOI: https://doi.org/10.1371/journal.ppat.1006474
- Primary Citation of Related Structures:
5WWP - PubMed Abstract:
Middle East respiratory syndrome coronavirus (MERS-CoV) remains a threat to public health worldwide; however, effective vaccine or drug against CoVs remains unavailable. CoV helicase is one of the three evolutionary most conserved proteins in nidoviruses, thus making it an important target for drug development. We report here the first structure of full-length coronavirus helicase, MERS-CoV nsp13. MERS-CoV helicase has multiple domains, including an N-terminal Cys/His rich domain (CH) with three zinc atoms, a beta-barrel domain and a C-terminal SF1 helicase core with two RecA-like subdomains. Our structural analyses show that while the domain organization of nsp13 is conserved throughout nidoviruses, the individual domains of nsp13 are closely related to the equivalent eukaryotic domains of Upf1 helicases. The most distinctive feature differentiating CoV helicases from eukaryotic Upf1 helicases is the interaction between CH domain and helicase core.
- MOH key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, No.9 Dong Dan San Tiao, Beijing, China.
Organizational Affiliation: