Split luciferase-based biosensors for characterizing EED binders
Li, L., Feng, L., Shi, M., Zeng, J., Chen, Z., Zhong, L., Huang, L., Guo, W., Huang, Y., Qi, W., Lu, C., Li, E., Zhao, K., Gu, J.(2017) Anal Biochem 522: 37-45
- PubMed: 28111304
- DOI: https://doi.org/10.1016/j.ab.2017.01.014
- Primary Citation of Related Structures:
5WUK - PubMed Abstract:
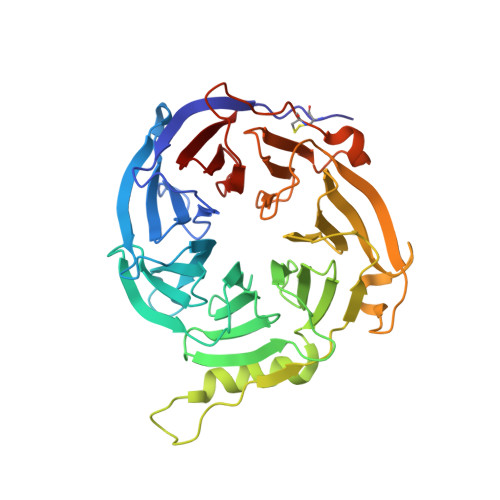
The EED (embryonic ectoderm development) subunit of the Polycomb repressive complex 2 (PRC2) plays an important role in the feed forward regulation of the PRC2 enzymatic activity. We recently identified a new class of allosteric PRC2 inhibitors that bind to the H3K27me3 pocket of EED. Multiple assays were developed and used to identify and characterize this type of PRC2 inhibitors. One of them is a genetically encoded EED biosensor based on the EED[G255D] mutant and the split firefly luciferase. This EED biosensor can detect the compound binding in the transfected cells and in the in vitro biochemical assays. Compared to other commonly used cellular assays, the EED biosensor assay has the advantage of shorter compound incubation with cells. The in vitro EED biosensor is much more sensitive than other label-free biophysical assays (e.g. DSF, ITC). Based on the crystal structure, the DSF data as well as the biosensor assay data, it's most likely that compound-induced increase in the luciferase activity of the EED[G255D] biosensor results from the decreased non-productive interactions between the EED subdomain and other subdomains within the biosensor construct. This new insight of the mechanism might help to broaden the use of the split luciferase based biosensors.
- China Novartis Institutes for Biomedical Research, 4218 Jinke Road, Shanghai 201203, China. Electronic address: ling-1.li@novartis.com.
Organizational Affiliation: