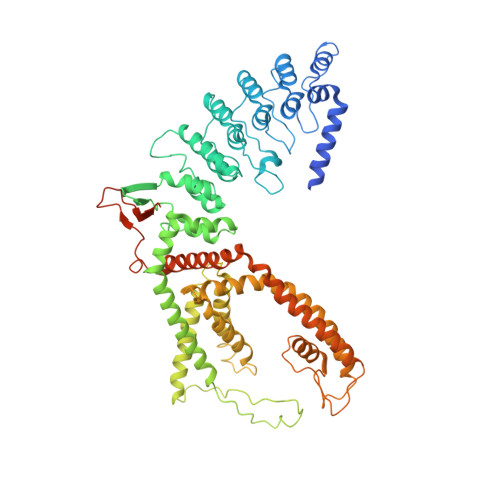
Swapping of transmembrane domains in the epithelial calcium channel TRPV6.
Singh, A.K., Saotome, K., Sobolevsky, A.I.(2017) Sci Rep 7: 10669-10669
- PubMed: 28878326
- DOI: https://doi.org/10.1038/s41598-017-10993-9
- Primary Citation of Related Structures:
5WO6, 5WO7, 5WO8, 5WO9, 5WOA - PubMed Abstract:
Tetrameric ion channels have either swapped or non-swapped arrangements of the S1-S4 and pore domains. Here we show that mutations in the transmembrane domain of TRPV6 can result in conversion from a domain-swapped to non-swapped fold. These results reveal structural determinants of domain swapping and raise the possibility that a single ion channel subtype can fold into either arrangement in vivo, affecting its function in normal or disease states.
- Department of Biochemistry and Molecular Biophysics, Columbia University 650 West 168th Street, New York, NY, 10032, USA.
Organizational Affiliation: