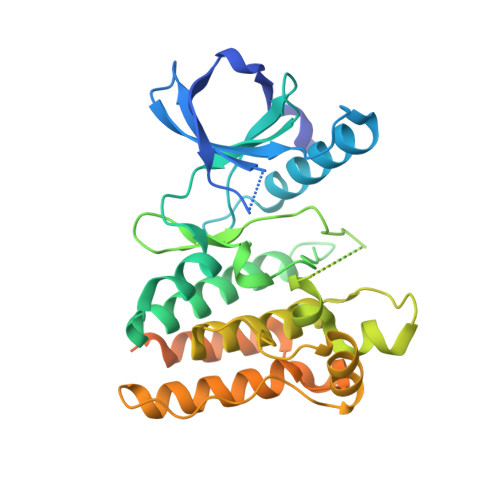
Crystal Structure of Ripk4 Reveals Dimerization-Dependent Kinase Activity.
Huang, C.S., Oberbeck, N., Hsiao, Y.C., Liu, P., Johnson, A.R., Dixit, V.M., Hymowitz, S.G.(2018) Structure 26: 767
- PubMed: 29706531
- DOI: https://doi.org/10.1016/j.str.2018.04.002
- Primary Citation of Related Structures:
5WNI, 5WNJ, 5WNK, 5WNL, 5WNM - PubMed Abstract:
Receptor-interacting protein kinase 4 (RIPK4) is a highly conserved regulator of epidermal differentiation. Members of the RIPK family possess a common kinase domain as well as unique accessory domains that likely dictate subcellular localization and substrate preferences. Mutations in human RIPK4 manifest as Bartsocas-Papas syndrome (BPS), a genetic disorder characterized by severe craniofacial and limb abnormalities. We describe the structure of the murine Ripk4 (MmRipk4) kinase domain, in ATP- and inhibitor-bound forms. The crystallographic dimer of MmRipk4 is similar to those of RIPK2 and BRAF, and we show that the intact dimeric entity is required for MmRipk4 catalytic activity through a series of engineered mutations and cell-based assays. We also assess the impact of BPS mutations on protein structure and activity to elucidate the molecular origins of the disease.
- Department of Structural Biology, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: