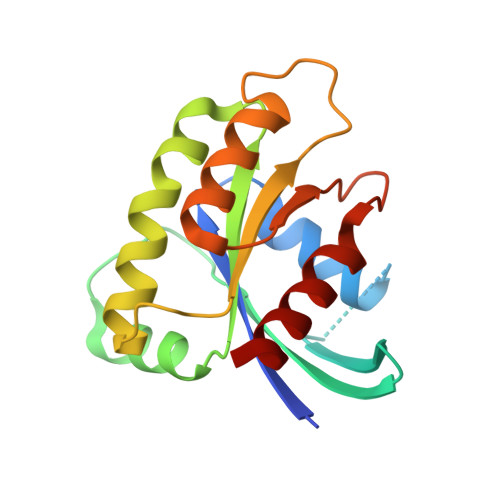
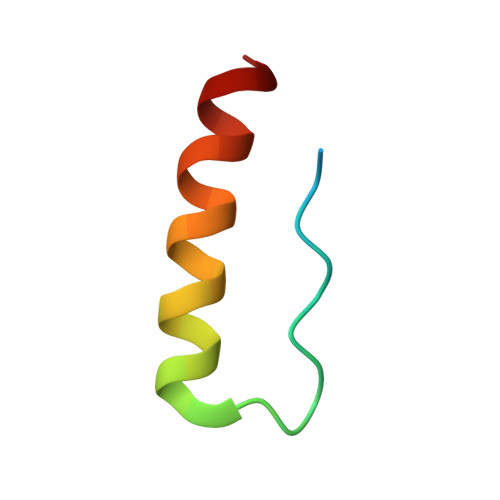
Exceptionally high-affinity Ras binders that remodel its effector domain.
McGee, J.H., Shim, S.Y., Lee, S.J., Swanson, P.K., Jiang, S.Y., Durney, M.A., Verdine, G.L.(2018) J Biological Chem 293: 3265-3280
- PubMed: 29282294
- DOI: https://doi.org/10.1074/jbc.M117.816348
- Primary Citation of Related Structures:
5WHA, 5WHB, 5WHD, 5WHE, 5WLB, 5WPL, 5WPM - PubMed Abstract:
The Ras proteins are aberrantly activated in a wide range of human cancers, often endowing tumors with aggressive properties and resistance to therapy. Decades of effort to develop direct Ras inhibitors for clinical use have thus far failed, largely because of a lack of adequate small-molecule-binding pockets on the Ras surface. Here, we report the discovery of Ras-binding miniproteins from a naïve library and their evolution to afford versions with midpicomolar affinity to Ras. A series of biochemical experiments indicated that these miniproteins bind to the Ras effector domain as dimers, and high-resolution crystal structures revealed that these miniprotein dimers bind Ras in an unprecedented mode in which the Ras effector domain is remodeled to expose an extended pocket that connects two isolated pockets previously found to engage small-molecule ligands. We also report a Ras point mutant that stabilizes the protein in the open conformation trapped by these miniproteins. These findings provide new tools for studying Ras structure and function and present opportunities for the development of both miniprotein and small-molecule inhibitors that directly target the Ras proteins.
- From the Departments of Molecular and Cellular Biology.
Organizational Affiliation: