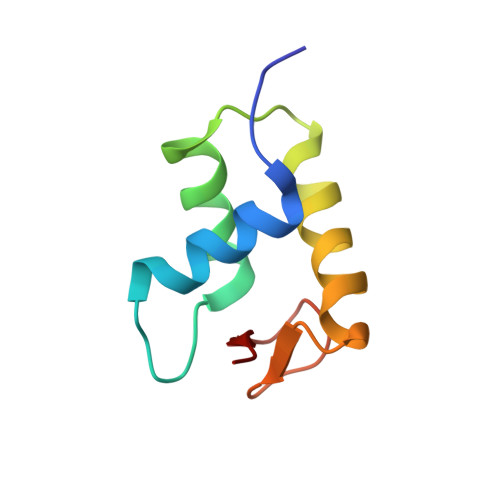
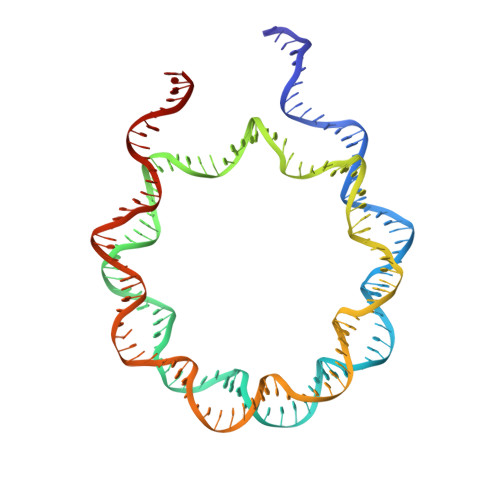
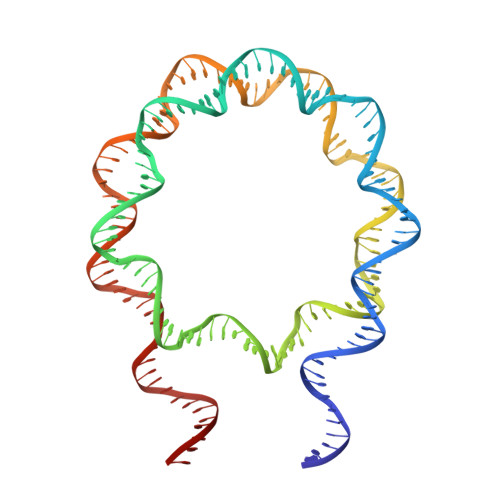
Revisit of Reconstituted 30-nm Nucleosome Arrays Reveals an Ensemble of Dynamic Structures.
Zhou, B.R., Jiang, J., Ghirlando, R., Norouzi, D., Sathish Yadav, K.N., Feng, H., Wang, R., Zhang, P., Zhurkin, V., Bai, Y.(2018) J Mol Biology 430: 3093-3110
- PubMed: 29959925
- DOI: https://doi.org/10.1016/j.jmb.2018.06.020
- Primary Citation of Related Structures:
5WCU - PubMed Abstract:
It has long been suggested that chromatin may form a fiber with a diameter of ~30 nm that suppresses transcription. Despite nearly four decades of study, the structural nature of the 30-nm chromatin fiber and conclusive evidence of its existence in vivo remain elusive. The key support for the existence of specific 30-nm chromatin fiber structures is based on the determination of the structures of reconstituted nucleosome arrays using X-ray crystallography and single-particle cryo-electron microscopy coupled with glutaraldehyde chemical cross-linking. Here we report the characterization of these nucleosome arrays in solution using analytical ultracentrifugation, NMR, and small-angle X-ray scattering. We found that the physical properties of these nucleosome arrays in solution are not consistent with formation of just a few discrete structures of nucleosome arrays. In addition, we obtained a crystal of the nucleosome in complex with the globular domain of linker histone H5 that shows a new form of nucleosome packing and suggests a plausible alternative compact conformation for nucleosome arrays. Taken together, our results challenge the key evidence for the existence of a limited number of structures of reconstituted nucleosome arrays in solution by revealing that the reconstituted nucleosome arrays are actually best described as an ensemble of various conformations with a zigzagged arrangement of nucleosomes. Our finding has implications for understanding the structure and function of chromatin in vivo.
- Laboratory of Biochemistry and Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: