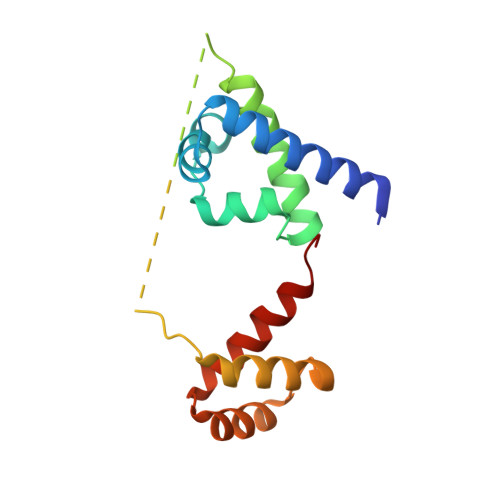
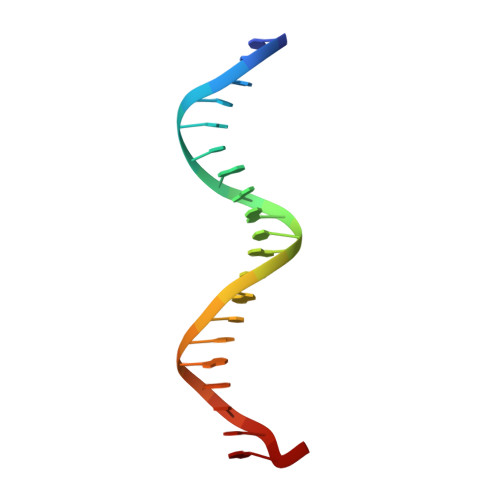
Biochemical and structural characterization of a novel cooperative binding mode by Pit-1 with CATT repeats in the macrophage migration inhibitory factor promoter.
Agarwal, S., Cho, T.Y.(2018) Nucleic Acids Res 46: 929-941
- PubMed: 29186613
- DOI: https://doi.org/10.1093/nar/gkx1183
- Primary Citation of Related Structures:
5WC9 - PubMed Abstract:
Overexpression of the proinflammatory cytokine macrophage migration inhibitory factor (MIF) is linked to a number of autoimmune diseases and cancer. MIF production has been correlated to the number of CATT repeats in a microsatellite region upstream of the MIF gene. We have characterized the interaction of pituitary-specific positive transcription factor 1 (Pit-1) with a portion of the MIF promoter region flanking a microsatellite polymorphism (-794 CATT5-8). Using fluorescence anisotropy, we quantified tight complex formation between Pit-1 and an oligonucleotide consisting of eight consecutive CATT repeats (8xCATT) with an apparent Kd of 35 nM. Using competition experiments we found a 23 base pair oligonucleotide with 4xCATT repeats to be the minimum DNA sequence necessary for high affinity interaction with Pit-1. The stoichiometry of the Pit-1 DNA interaction was determined to be 2:1 and binding is cooperative in nature. We subsequently structurally characterized the complex and discovered a completely novel binding mode for Pit-1 in contrast to previously described Pit-1 complex structures. The affinity of Pit-1 for the CATT target sequence was found to be highly dependent on cooperativity. This work lays the groundwork for understanding transcriptional regulation of MIF and pursuing Pit-1 as a therapeutic target to treat MIF-mediated inflammatory disorders.
- Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, St. Louis, MO 63104, USA.
Organizational Affiliation: