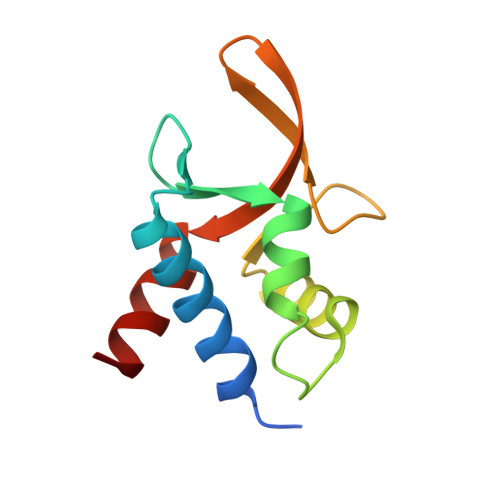
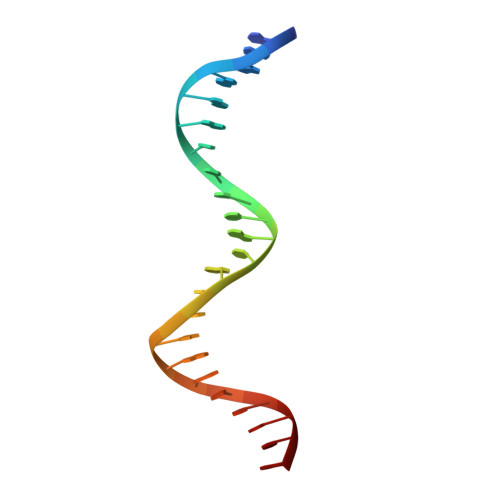
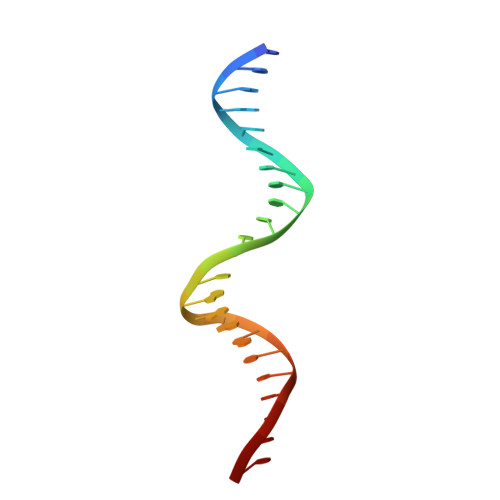
Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Traore, D.A.K., Wisniewski, J.A., Flanigan, S.F., Conroy, P.J., Panjikar, S., Mok, Y.F., Lao, C., Griffin, M.D.W., Adams, V., Rood, J.I., Whisstock, J.C.(2018) Nat Commun 9: 3732-3732
- PubMed: 30213934
- DOI: https://doi.org/10.1038/s41467-018-06096-2
- Primary Citation of Related Structures:
5VFX, 5VFY - PubMed Abstract:
Conjugation is fundamental for the acquisition of new genetic traits and the development of antibiotic resistance in pathogenic organisms. Here, we show that a hypothetical Clostridium perfringens protein, TcpK, which is encoded by the tetracycline resistance plasmid pCW3, is essential for efficient conjugative DNA transfer. Our studies reveal that TcpK is a member of the winged helix-turn-helix (wHTH) transcription factor superfamily and that it forms a dimer in solution. Furthermore, TcpK specifically binds to a nine-nucleotide sequence that is present as tandem repeats within the pCW3 origin of transfer (oriT). The X-ray crystal structure of the TcpK-TcpK box complex reveals a binding mode centered on and around the β-wing, which is different from what has been previously shown for other wHTH proteins. Structure-guided mutagenesis experiments validate the specific interaction between TcpK and the DNA molecule. Additional studies highlight that the TcpK dimer is important for specific DNA binding.
- Department of Biochemistry and Molecular Biology, Infection and Immunity Program, Monash Biomedicine Discovery Institute, Monash University, Clayton, 3800, VIC, Australia.
Organizational Affiliation: