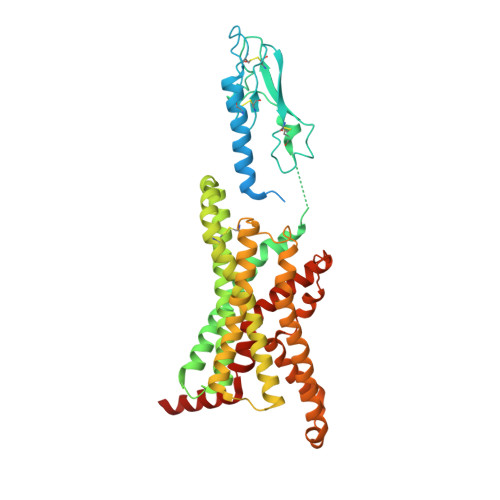
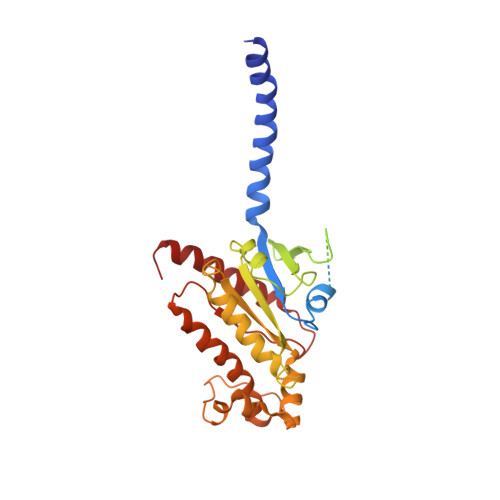
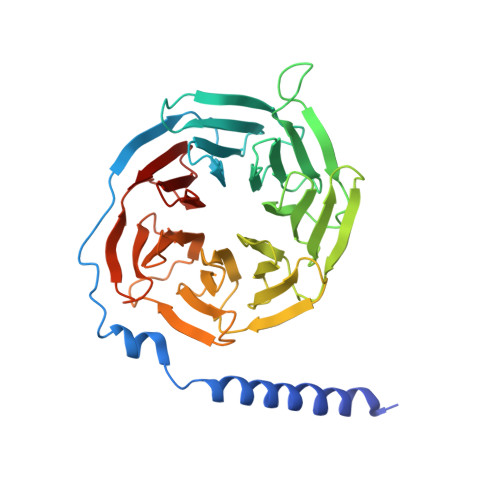
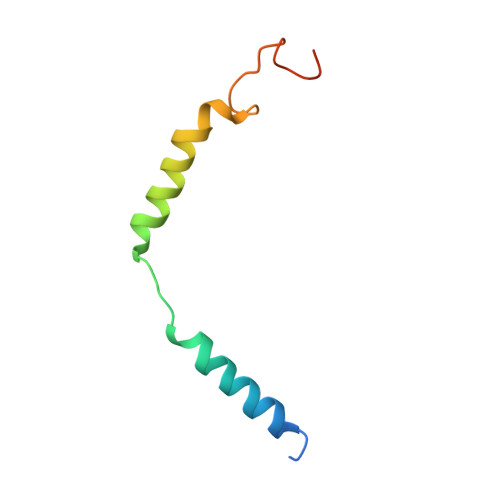
Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Zhang, Y., Sun, B., Feng, D., Hu, H., Chu, M., Qu, Q., Tarrasch, J.T., Li, S., Sun Kobilka, T., Kobilka, B.K., Skiniotis, G.(2017) Nature 546: 248-253
- PubMed: 28538729
- DOI: https://doi.org/10.1038/nature22394
- Primary Citation of Related Structures:
5VAI - PubMed Abstract:
Glucagon-like peptide 1 (GLP-1) is a hormone with essential roles in regulating insulin secretion, carbohydrate metabolism and appetite. GLP-1 effects are mediated through binding to the GLP-1 receptor (GLP-1R), a class B G-protein-coupled receptor (GPCR) that signals primarily through the stimulatory G protein G s . Class B GPCRs are important therapeutic targets; however, our understanding of their mechanism of action is limited by the lack of structural information on activated and full-length receptors. Here we report the cryo-electron microscopy structure of the peptide-activated GLP-1R-G s complex at near atomic resolution. The peptide is clasped between the N-terminal domain and the transmembrane core of the receptor, and further stabilized by extracellular loops. Conformational changes in the transmembrane domain result in a sharp kink in the middle of transmembrane helix 6, which pivots its intracellular half outward to accommodate the α5-helix of the Ras-like domain of G s . These results provide a structural framework for understanding class B GPCR activation through hormone binding.
- Life Sciences Institute and Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, Michigan 48109, USA.
Organizational Affiliation: