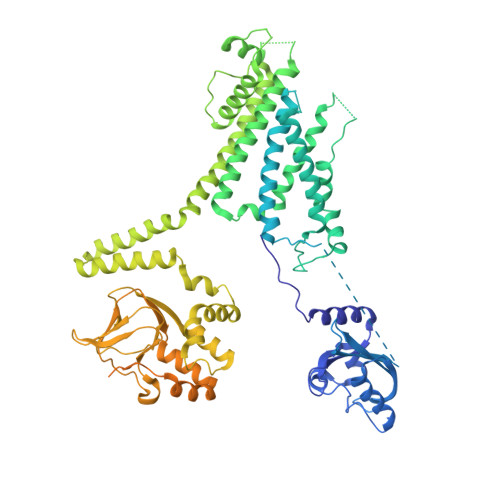
Cryo-EM Structure of the Open Human Ether-a-go-go-Related K(+) Channel hERG.
Wang, W., MacKinnon, R.(2017) Cell 169: 422-430.e10
- PubMed: 28431243
- DOI: https://doi.org/10.1016/j.cell.2017.03.048
- Primary Citation of Related Structures:
5VA1, 5VA2, 5VA3 - PubMed Abstract:
The human ether-à-go-go-related potassium channel (hERG, Kv11.1) is a voltage-dependent channel known for its role in repolarizing the cardiac action potential. hERG alteration by mutation or pharmacological inhibition produces Long QT syndrome and the lethal cardiac arrhythmia torsade de pointes. We have determined the molecular structure of hERG to 3.8 Å using cryo-electron microscopy. In this structure, the voltage sensors adopt a depolarized conformation, and the pore is open. The central cavity has an atypically small central volume surrounded by four deep hydrophobic pockets, which may explain hERG's unusual sensitivity to many drugs. A subtle structural feature of the hERG selectivity filter might correlate with its fast inactivation rate, which is key to hERG's role in cardiac action potential repolarization.
- Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University and Howard Hughes Medical Institute, 1230 York Avenue, New York, NY 10065, USA.
Organizational Affiliation: