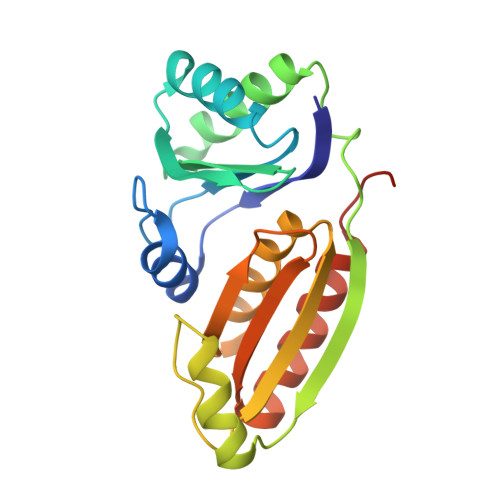
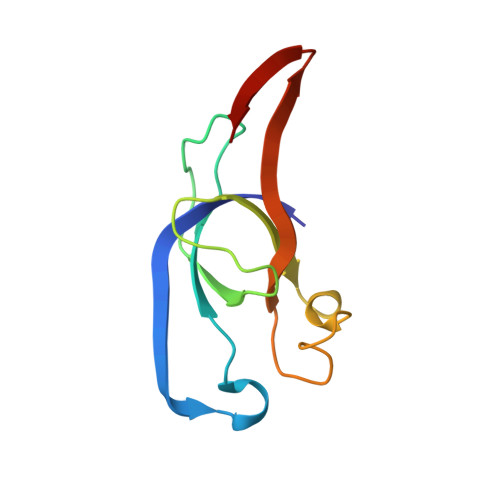Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell.
Sutter, M., Greber, B., Aussignargues, C., Kerfeld, C.A.(2017) Science 356: 1293-1297
- PubMed: 28642439
- DOI: https://doi.org/10.1126/science.aan3289
- Primary Citation of Related Structures:
5V74, 5V75, 5V76 - PubMed Abstract:
Many bacteria contain primitive organelles composed entirely of protein. These bacterial microcompartments share a common architecture of an enzymatic core encapsulated in a selectively permeable protein shell; prominent examples include the carboxysome for CO 2 fixation and catabolic microcompartments found in many pathogenic microbes. The shell sequesters enzymatic reactions from the cytosol, analogous to the lipid-based membrane of eukaryotic organelles. Despite available structural information for single building blocks, the principles of shell assembly have remained elusive. We present the crystal structure of an intact shell from Haliangium ochraceum , revealing the basic principles of bacterial microcompartment shell construction. Given the conservation among shell proteins of all bacterial microcompartments, these principles apply to functionally diverse organelles and can inform the design and engineering of shells with new functionalities.
- Michigan State University-U.S. Department of Energy (MSU-DOE) Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA.
Organizational Affiliation: