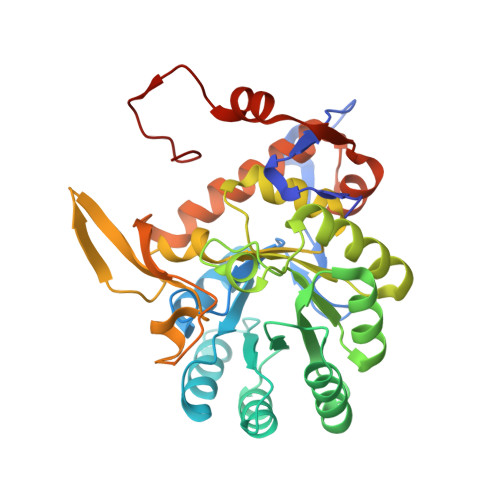
Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P182
Maltseva, N., Kim, Y., Mulligan, R., Makowska-Grzyska, M., Gu, M., Gollapalli, D.R., Hedstrom, L., Joachimiak, A., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.